tage Overview
BioGears® simulation UI source code is hosted here. Our latest deployment is still in a beta phase, and is intended to be an intermediate release to showcase the capabilities of the BioGears® simulation UI. This version of the software is meant to elicit feedback and enhance community involvement in establishing end product expectations.
| Platform | Compiler | Architecture | Status |
|---|---|---|---|
| Windows | msvc15 | amd64 | 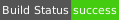 |
| Windows | msvc16 | amd64 | 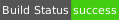 |
| Linux | gcc9 | amd64 | 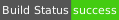 |
| MacOS Yosemite | clang10 | amd64 | 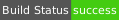 |
| MacOS Catalina | clang11 | amd64 |  |
- To allow users to visually compose and execute BioGears® simulation scenarios
- To allow users to visualize BioGears® scenario actions and outputs in an intuitive timeline format
BioGears® simulation UI requires the following dependencies:
- BioGears Physiology Engine - build requirements can be found here.
- Boost - build requirements can be found here.
- QT 5 - build requirements can be found here.
A submodule for BioGears is located in projects/libbiogears. It can be initialized using the git submodule commands from the root level.
git submodule init projects/libbiogears
git submodule update
BioGears should then be built either inside the submodule or at the root level in its own build folder for example
mkdir build-biogears
cd build-biogears
cmake ../projectslibbiogears -DCMAKE_INSTALL_PREFIX=${PWD}../build-gui/usr
cmake --build . -config Release -target install
If you are going to build BioGears only once it is fine to place BioGears inside the same folder structure as its deps, but if you are planning on building BioGears multiple times and testing the different versions of BioGears against the GUI then the example above places the BioGears libraries in its own external tree so that the build system for biogears will never see the headers for a previously installed version.
You can follow the instructions for building biogears from its github page for more information, just remember to account of the unique structure layout
#Pulling Code
git clone https://github.com/BioGearsEngine/ui.git visualizer
cd visualizer
git submodule init
git submodule update --progress
#Building the Visualizer code against biogears
cd ..
mkdir build-ui
cd build-ui
cmake .. -G Ninja -DCMAKE_PREFIX_PATH="/opt/biogears/external/"
cmake -DCMAKE_BUILD_TYPE=Release .
cmake --build . --config Release --target install
cmake --build . --config Release --target gather_runtime_dependencies
Further instructions can be found in our WIKI at our github page
Once open in Microsoft Visual Studios (MVS), right-click on BioGearsUI on the lefthand side of the screen in the Solution Explorer. Switch the configuration in the top left corner of the property page to be "All Configurations." On the left hand side of the screen, navigate to Configuration Properties→Debugging, and fill in the Environment box with the QT Path:
- QT_PLUGIN_PATH=D:\remotes\sed-stash\biogears\external\windows-vc15-amd64\plugins
Note: The above path is an example. Be sure to switch the path to your specific plugin folder of the windows build based on your folder configuration.
Specify the configuration (release or debug) you wish to build in at the top of the MVS screen. Click the F7 key or navigate to the Build→Build Solution tab at the top of the screen. Make sure the build is successful. Then, navigate in the solution explorer to CMakePredefinedTargets→gather_runtime_dependencies, right-clock on stage, and select build. Make sure this build is also successful.
Click F5 or click the play button to run the solution. The viualizer/UI should pop up and prompt you with further directions.
Note: For full details, visit our wiki here.
QT Creator supports CMake project imports. The following is a short guide on how to setup a project with QT Creator
##Code of Conduct
We support the contributor covenant and the scope and enforcement it details. We reserve the right of enforcement that we feel is appropriate given the nature of the offense up to and including a permanent ban from the project.
##Contributing
Details will be filled in shortly. In the meantime if you have a contribution or issue to post/push feel free to contact biogears@ara.com with the details.
##Additional Documentation
For more detailed documentation including model discussions and implementation details can be found at www.BioGearsEngine.com