OpenFoam .org version 9 has been used to run the simulation in this repository. I would recommend installing OpenFOAM V9 through docker, following the guide at https://openfoam.org/download/9-linux/. For linux this leads to a straight-forward setup, which is easily accessible. There are no instructions on Windows, but this is fairly easy.:
- Download and install Docker: https://docs.docker.com/desktop/windows/install/. You might need to setup WSL.
- Once setup, go to https://hub.docker.com/r/openfoam/openfoam9-paraview56 and use the docker pull command to start installing the docker.
- Run the docker and access it through a terminal, set up some-sort of shared folder in order to work efficiently.
After an installation like this, you access the docker image first and then execute OpenFOAM specific commands.
All commands use a certain naming convention, so they are easy to find with autocompletion.
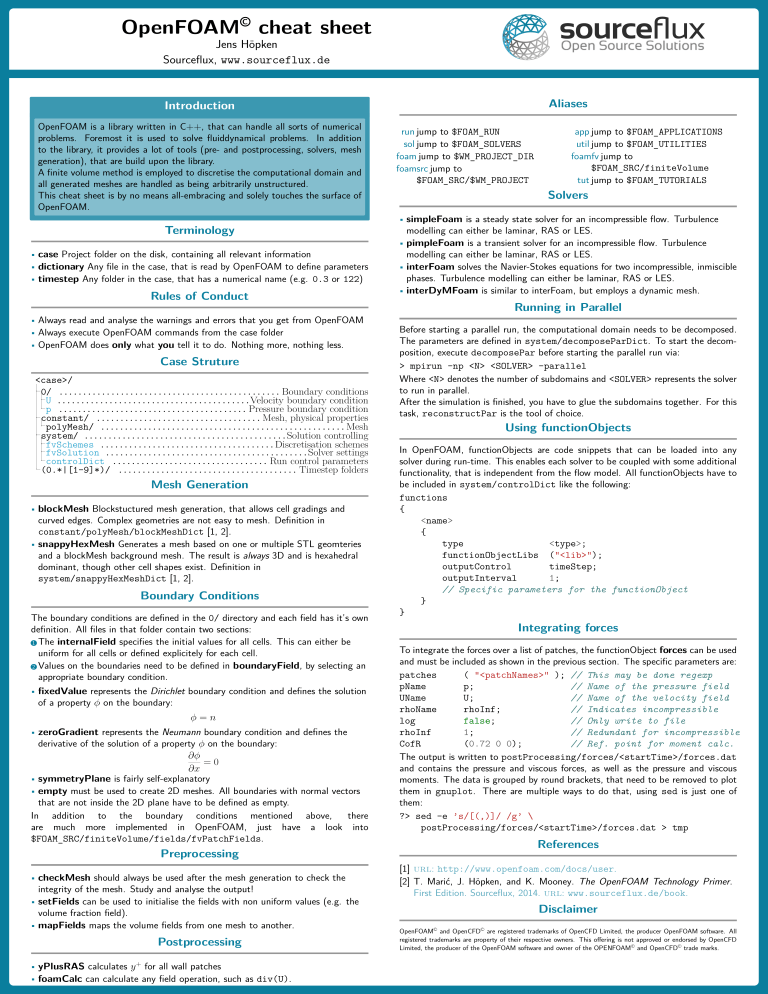
To give you a head-start, see the cheat-sheet below.
Solver capability matrix (Note: for openfoam.com version, mostly the same but might differ!)
PyFoam Very convenient library which adds functionality and tools for OpenFoam
- Used to monitor every simulation with: pyFoamPlotWatcher --with-all log
- Opens number of real-time plots to monitor residuals, specific values
- customRegExp found under some simulation is also shown every timestep, in this case it calculates the total average gas concentration [ppm] using functionObjects
Paraview has been used for analysis of simulation results. A post-processing script is provided to automatically apply the results of the DAEI
- Installation instructions can be found on the website
- Make sure that you install a version of paraview with python! Otherwise the macro-script will not work.
First this script needs to be imported into ParaView:
- To get the script: install this library and run
AEI CopyExamples, the scripty AEI.py copied in your active directory is the ParaView macro. - Or download it here from the git manually Once the DAEI script is imported:
- Import the script it in ParaView by copying it to the macro folder, or through the macro toolbar option.
- Select the source-simulation (.foam file in the source-tree)
- Make sure that there is a AEI_Norm.csv available in the ParaView source-tree, values are directly read and applied from there.
- Run the script AEI.py, A pop-up will open
- give the text-box a comma-separated list of chemicals, such as: "C10H16","CO2","O3"
- press ok/apply/enter, almost instantly many calculators will be added to the source-tree. The final result is put into the AEI calculator, all the other ones are appropriately named.
To run the steps a few things need to be prepared, all the following steps need to be run in an installation where OpenFOAM commands are available. So in the docker, or load your environment variables.
-
Mesh files describing the geometry, located in
<case>/constant/geometry/*.stlor originating from<case>/system/blockMeshDict- in the case of
*.stlfiles you have to run do a few things:- run
blockMeshto generate the base block, that needs to bigger then the geometry. - run
surfaceFeaturesto extract surface features from the files. - run
snappyHexMesh -overwriteto generate the final geometry, this might take a while.- In some cases snappyHexMesh generates folders like
<case>/0.001/and<case>/0.002/, copy the folder polyMesh to constant and the other files to the<case>/0/folder
cd <case>/ blockMesh | tee log.blockMesh surfaceFeatures | tee log.surfaceFeatures snappyHexMesh | tee log.snappyHexMesh cp -r 0.002/polyMesh constant/polyMesh cp 0.002/*Level 0/ - In some cases snappyHexMesh generates folders like
- run
- in the case of
blockMeshDictsimulations simply runblockMesh
Now for sanity, we want to run the simulation a bit more optimized and use all our processors in parallel.
- in the case of
- Edit
<case>/system/decomposeParDict- Change the line
numberOfSubdomains 16;to the amount of physical cores your processor has (not threads). You can check this in task-manager on Windows.
- Change the line
- Now run
decomposeParin the<case>/folder, this will split the simulation in subdomains.
As a last step you can check if everything works with Paraview:
- make a file with
.foamextension in the<case>/directory and open it with openfoam:touch <case>/simulation.foam paraview <case>/simulation.foam & - You can also open the case through the paraview interface.
- You can inspect the generated geometry, and see how
decomposeParsplit the problem in sub-domains. By using the wireframe or Surface with edges visualization, the mesh can be visualized.
Most cases here use the buoyantReactingFoam solver, so this general description is written for those cases.
- Go to the
<case>/directory in your openFOAM environment - run
mpirun -np <No_Cores> buoyantReactingFoam -parallel | tee log- where
<No_Cores>is thenumberOfSubdomainsyou set before in<case>/system/decomposeParDict
cd <case>/ mpirun -np 16 buoyantReactingFoam -parallel | tee log - where
With PyFoam you can monitor the simulation visually by graphing the log in realtime.
- first you need to install it on your system, so not in the docker with the command
pip install PyFoamor your package manager of choice for python. pyFoamPlotWatcher --with-all log- Now a bunch of plots pop-up, among others: residuals, some concentrations changing over time (
customRegExp), courant number, etc. - While running, you can open the
*.foamfile with Paraview. Set the case to decomposed and you can view any new timesteps after refreshing. - Stop the simulation with CTRL+C in the command-line window.
- If you want a nicer view, and the decomposed case to be restored:
reconstructParto reconstruct all timestepsreconstructPar --latestTimeto only reconstruct the last available timestep.