diff --git a/.github/workflows/CI.yml b/.github/workflows/CI.yml
index 7b3c420..49acf7b 100644
--- a/.github/workflows/CI.yml
+++ b/.github/workflows/CI.yml
@@ -2,7 +2,7 @@ name: CI
on:
push:
- branches: [ "main", "docs", "patch", "feature" ]
+ branches: [ "main", "docs", "patch", "feature", "nodrawci" ]
pull_request:
branches: [ "main" ]
diff --git a/.github/workflows/nb.yml b/.github/workflows/nb.yml
index 81ba8e1..2b22441 100644
--- a/.github/workflows/nb.yml
+++ b/.github/workflows/nb.yml
@@ -2,7 +2,7 @@ name: Notebooks
on:
push:
- branches: [ "main", "patch", "feature" ]
+ branches: [ "main", "patch", "feature", "nodrawci" ]
pull_request:
branches: [ "main" ]
@@ -13,7 +13,7 @@ jobs:
strategy:
fail-fast: false
matrix:
- python-version: ['3.9']
+ python-version: ['3.10']
steps:
- uses: actions/checkout@v3
- name: Set up Python ${{ matrix.python-version }}
diff --git a/README.md b/README.md
index 16586f6..94bcf5e 100644
--- a/README.md
+++ b/README.md
@@ -98,13 +98,12 @@ df.head(3)
| Column | Description | Required |
|:----------|:------------------------------------------------|:----------|
- | `var` | Variable field | |
+ | `var` | Variable label | ✓ |
| `r` | Correlation coefficients (estimates to plot) | ✓ |
- | `moerror` | Conf. int.'s *margin of error*. | |
| `label` | Variable labels | ✓ |
| `group` | Variable grouping labels | |
- | `ll` | Conf. int. *lower limits* | ✓ |
- | `hl` | Containing the conf. int. *higher limits* | ✓ |
+ | `ll` | Conf. int. *lower limits* | |
+ | `hl` | Containing the conf. int. *higher limits* | |
| `n` | Sample size | |
| `power` | Statistical power | |
| `p-val` | P-value | |
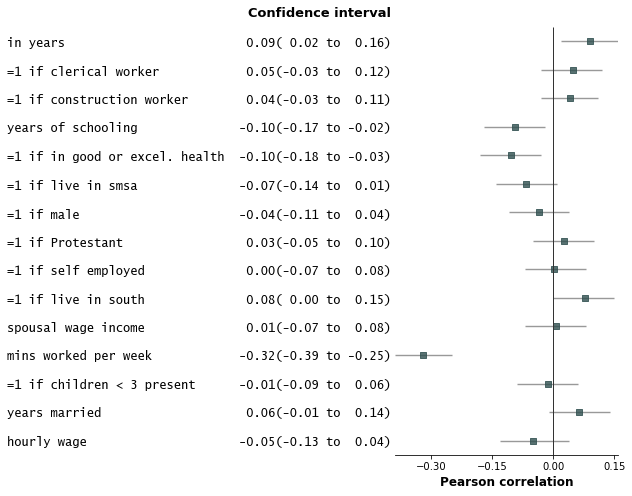
@@ -122,7 +121,7 @@ fp.forestplot(df, # the dataframe with results data
xlabel="Pearson correlation", # x-label title
)
```
-
+
Save the plot
```python
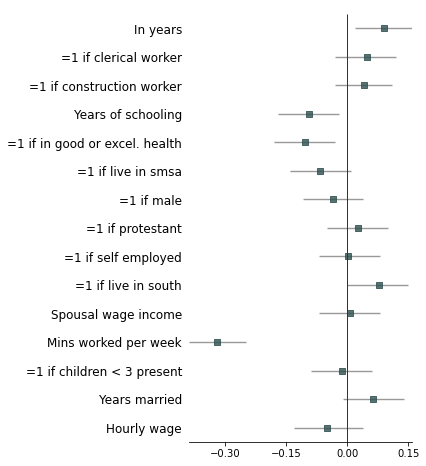
@@ -202,7 +201,7 @@ fp.forestplot(df, # the dataframe with results data
**{'fontfamily': 'sans-serif'} # revert to sans-serif
)
```
-
+
5. Example with more customizations
```python
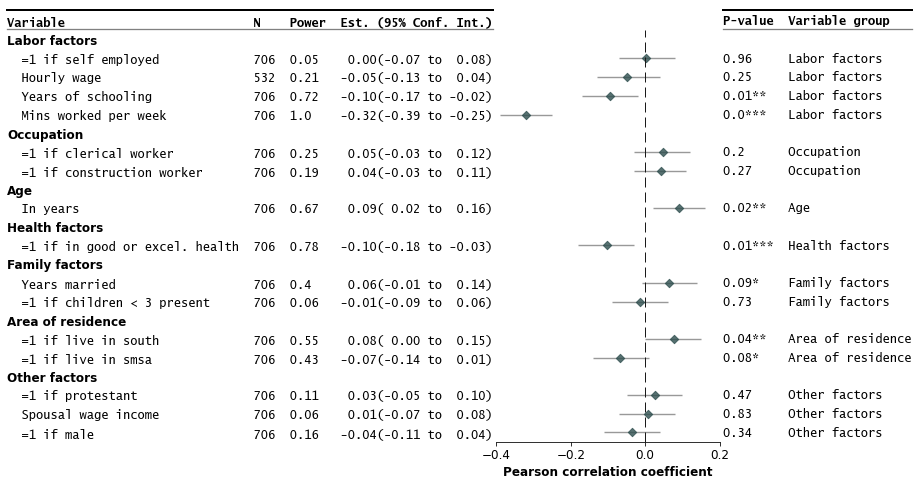
@@ -232,7 +231,7 @@ fp.forestplot(df, # the dataframe with results data
}
)
```
-
+
Annotations arguments allowed include:
@@ -259,8 +258,6 @@ fp.forestplot(df, # the dataframe with results data
(back to top)
-
-
## Gallery and API Options[](#gallery-and-api-options)
@@ -276,8 +273,8 @@ More fined-grained control for base plot options (eg font sizes, marker colors)
| `dataframe` | Pandas dataframe where rows are variables (or studies for meta-analyses) and columns include estimated effect sizes, labels, and confidence intervals, etc. | ✓ |
| `estimate` | Name of column in `dataframe` containing the *estimates*. | ✓ |
| `varlabel` | Name of column in `dataframe` containing the *variable labels* (study labels if meta-analyses). | ✓ |
-| `ll` | Name of column in `dataframe` containing the conf. int. *lower limits*. | ✓ |
-| `hl` | Name of column in `dataframe` containing the conf. int. *higher limits*. | ✓ |
+| `ll` | Name of column in `dataframe` containing the conf. int. *lower limits*. | |
+| `hl` | Name of column in `dataframe` containing the conf. int. *higher limits*. | |
| `logscale` | If True, make the x-axis log scale. Default is False. | |
| `capitalize` | How to capitalize strings. Default is None. One of "capitalize", "title", "lower", "upper", "swapcase". | |
| `form_ci_report` | If True (default), report the estimates and confidence interval beside the variable labels. | |
diff --git a/forestplot/__init__.py b/forestplot/__init__.py
index f5bed59..cb4613b 100644
--- a/forestplot/__init__.py
+++ b/forestplot/__init__.py
@@ -1,5 +1,5 @@
"""State version and import user-facing functions."""
-VERSION = (0, 2, 2)
+VERSION = (0, 3, 0)
__version__ = ".".join(map(str, VERSION))
diff --git a/forestplot/arg_validators.py b/forestplot/arg_validators.py
index efddb92..1d22df5 100644
--- a/forestplot/arg_validators.py
+++ b/forestplot/arg_validators.py
@@ -14,6 +14,7 @@ def check_data(
group_order: Optional[Sequence] = None,
ll: Optional[str] = None,
hl: Optional[str] = None,
+ form_ci_report: bool = None,
annote: Optional[Union[Sequence[str], None]] = None,
annoteheaders: Optional[Union[Sequence[str], None]] = None,
rightannote: Optional[Union[Sequence[str], None]] = None,
@@ -43,6 +44,8 @@ def check_data(
Name of column containing the lower limit of the confidence intervals.
hl (str)
Name of column containing the upper limit of the confidence intervals.
+ form_ci_report (bool)
+ If True, form the formatted confidence interval as a string.
annote (list-like)
List of columns to add as additional annotation in the plot.
annoteheaders (list-like)
@@ -61,6 +64,24 @@ def check_data(
-------
pd.core.frame.DataFrame.
"""
+ ##########################################################################
+ ## Check that CI options (ll, hl, form_ci_report) are consistent
+ ##########################################################################
+ if ll is None:
+ try:
+ assert hl is None
+ except Exception:
+ raise TypeError("'ll' is None. 'hl' should also be None.")
+
+ if hl is None:
+ try:
+ assert ll is None
+ except Exception:
+ raise TypeError("'hl' is None. 'll' should also be None.")
+
+ if ll is None and form_ci_report:
+ warnings.warn("'ll' is None. 'form_ci_report' will be set to False.")
+
##########################################################################
## Check that numeric data are numeric
##########################################################################
diff --git a/forestplot/graph_utils.py b/forestplot/graph_utils.py
index 6d31df3..3a7090a 100644
--- a/forestplot/graph_utils.py
+++ b/forestplot/graph_utils.py
@@ -43,17 +43,18 @@ def draw_ci(
-------
Matplotlib Axes object.
"""
- lw = kwargs.get("lw", 1.4)
- linecolor = kwargs.get("linecolor", ".6")
- ax.errorbar(
- x=dataframe[estimate],
- y=dataframe[yticklabel],
- xerr=[dataframe[estimate] - dataframe[ll], dataframe[hl] - dataframe[estimate]],
- ecolor=linecolor,
- elinewidth=lw,
- ls="none",
- zorder=0,
- )
+ if ll is not None:
+ lw = kwargs.get("lw", 1.4)
+ linecolor = kwargs.get("linecolor", ".6")
+ ax.errorbar(
+ x=dataframe[estimate],
+ y=dataframe[yticklabel],
+ xerr=[dataframe[estimate] - dataframe[ll], dataframe[hl] - dataframe[estimate]],
+ ecolor=linecolor,
+ elinewidth=lw,
+ ls="none",
+ zorder=0,
+ )
if logscale:
ax.set_xscale("log", base=10)
return ax
@@ -532,6 +533,7 @@ def format_xlabel(xlabel: str, ax: Axes, **kwargs: Any) -> Axes:
def format_xticks(
dataframe: pd.core.frame.DataFrame,
+ estimate: str,
ll: str,
hl: str,
xticks: Optional[Union[list, range]],
@@ -550,6 +552,9 @@ def format_xticks(
dataframe (pandas.core.frame.DataFrame)
Pandas DataFrame where rows are variables. Columns are variable name, estimates,
margin of error, etc.
+ estimate (str)
+ Name of column containing the estimates (e.g. pearson correlation coefficient,
+ OR, regression estimates, etc.).
ll (str)
Name of column containing the lower limit of the confidence intervals.
Optional
@@ -568,8 +573,12 @@ def format_xticks(
nticks = kwargs.get("nticks", 5)
xtick_size = kwargs.get("xtick_size", 10)
xticklabels = kwargs.get("xticklabels", None)
- xlowerlimit = dataframe[ll].min()
- xupperlimit = dataframe[hl].max()
+ if ll is not None:
+ xlowerlimit = dataframe[ll].min()
+ xupperlimit = dataframe[hl].max()
+ else:
+ xlowerlimit = 1.1 * dataframe[estimate].min()
+ xupperlimit = 1.1 * dataframe[estimate].max()
ax.set_xlim(xlowerlimit, xupperlimit)
if xticks is not None:
ax.set_xticks(xticks)
@@ -653,6 +662,7 @@ def draw_tablelines(
pval: str,
right_annoteheaders: Optional[Union[Sequence[str], None]],
ax: Axes,
+ **kwargs: Any
) -> Axes:
"""
Plot horizontal lines as table lines.
@@ -684,7 +694,7 @@ def draw_tablelines(
[x0, x1], [nrows - 1.45, nrows - 1.45], color="0.5", linewidth=lower_lw, clip_on=False
)
if (right_annoteheaders is not None) or (pval is not None):
- extrapad = 0.05
+ extrapad = kwargs.get("extrapad", 0.05)
x0 = ax.get_xlim()[1] * (1 + extrapad)
plt.plot(
[x0, righttext_width],
diff --git a/forestplot/plot.py b/forestplot/plot.py
index 1ec8f06..9c8b191 100644
--- a/forestplot/plot.py
+++ b/forestplot/plot.py
@@ -100,7 +100,7 @@ def forestplot(
form_ci_report (bool)
If True, form the formatted confidence interval as a string.
ci_report (bool)
- If True, form the formatted confidence interval as a string.
+ If True, report the formatted confidence interval as a string.
groupvar (str)
Name of column containing group of variables.
group_order (list-like)
@@ -170,8 +170,8 @@ def forestplot(
rightannote=rightannote,
right_annoteheaders=right_annoteheaders,
)
- if (ll is None) or (hl is None):
- ll, hl = "ll", "hl"
+ if ll is None:
+ ci_report = False
if ci_report is True:
form_ci_report = True
if preprocess:
@@ -371,7 +371,9 @@ def _make_forestplot(
draw_est_markers(
dataframe=dataframe, estimate=estimate, yticklabel=yticklabel, ax=ax, **kwargs
)
- format_xticks(dataframe=dataframe, ll=ll, hl=hl, xticks=xticks, ax=ax, **kwargs)
+ format_xticks(
+ dataframe=dataframe, estimate=estimate, ll=ll, hl=hl, xticks=xticks, ax=ax, **kwargs
+ )
draw_ref_xline(
ax=ax,
dataframe=dataframe,
diff --git a/forestplot/text_utils.py b/forestplot/text_utils.py
index c5d0025..b56aee5 100644
--- a/forestplot/text_utils.py
+++ b/forestplot/text_utils.py
@@ -49,16 +49,21 @@ def form_est_ci(
-------
pd.core.frame.DataFrame with an additional formatted 'est_ci' column.
"""
- for col in [estimate, ll, hl]:
+ if ll is None:
+ cols = [estimate]
+ else:
+ cols = [estimate, ll, hl]
+ for col in cols:
dataframe = _right_justify_num(
dataframe=dataframe, col=col, decimal_precision=decimal_precision
)
for ix, row in dataframe.iterrows():
formatted_est = row[f"formatted_{estimate}"]
- formatted_ll, formatted_hl = row[f"formatted_{ll}"], row[f"formatted_{hl}"]
- formatted_ci = "".join([caps[0], formatted_ll, connector, formatted_hl, caps[1]])
- dataframe.loc[ix, "ci_range"] = formatted_ci
- dataframe.loc[ix, "est_ci"] = "".join([formatted_est, formatted_ci])
+ if ll is not None:
+ formatted_ll, formatted_hl = row[f"formatted_{ll}"], row[f"formatted_{hl}"]
+ formatted_ci = "".join([caps[0], formatted_ll, connector, formatted_hl, caps[1]])
+ dataframe.loc[ix, "ci_range"] = formatted_ci
+ dataframe.loc[ix, "est_ci"] = "".join([formatted_est, formatted_ci])
return dataframe
diff --git a/setup.py b/setup.py
index 933e6ea..3088c5f 100644
--- a/setup.py
+++ b/setup.py
@@ -10,7 +10,7 @@
install_requires = ["pandas", "numpy", "matplotlib", "matplotlib-inline<=0.1.3"]
setup(
name="forestplot",
- version="0.2.2",
+ version="0.3.0",
license="MIT",
author="Lucas Shen",
author_email="lucas@lucasshen.com",
diff --git a/tests/test_arg_validators.py b/tests/test_arg_validators.py
index dcd585b..fde5653 100644
--- a/tests/test_arg_validators.py
+++ b/tests/test_arg_validators.py
@@ -25,9 +25,9 @@ def test_check_data():
check_data(dataframe=_df, estimate="estimate", varlabel="varlabel")
# Assert that assertion for numeric type for ll works
- _df = pd.DataFrame({"estimate": numeric, "ll": string})
+ _df = pd.DataFrame({"estimate": numeric, "ll": string, "hl": numeric})
with pytest.raises(TypeError) as excinfo:
- check_data(dataframe=_df, estimate="estimate", varlabel="estimate", ll="ll")
+ check_data(dataframe=_df, estimate="estimate", varlabel="estimate", ll="ll", hl="hl")
assert str(excinfo.value) == "CI lowerlimit values should be float or int"
# Assert that conversion for numeric ll stored as string works
@@ -41,9 +41,9 @@ def test_check_data():
check_data(dataframe=_df, estimate="estimate", varlabel="estimate", ll="ll", hl="hl")
# Assert that assertion for numeric type for hl works
- _df = pd.DataFrame({"estimate": numeric, "hl": string})
+ _df = pd.DataFrame({"estimate": numeric, "ll": numeric, "hl": string})
with pytest.raises(TypeError) as excinfo:
- check_data(dataframe=_df, estimate="estimate", varlabel="estimate", hl="hl")
+ check_data(dataframe=_df, estimate="estimate", varlabel="estimate", ll="ll", hl="hl")
assert str(excinfo.value) == "CI higherlimit values should be float or int"
# Assert that conversion for numeric hl stored as string works
@@ -56,6 +56,17 @@ def test_check_data():
)
check_data(dataframe=_df, estimate="estimate", varlabel="estimate", ll="ll", hl="hl")
+ # Assert that check for CI options are consistent works
+ _df = pd.DataFrame({"estimate": numeric, "ll": string, "hl": string})
+ with pytest.raises(TypeError) as excinfo:
+ check_data(dataframe=_df, estimate="estimate", varlabel="estimate", ll=None, hl="hl")
+ assert str(excinfo.value) == "'ll' is None. 'hl' should also be None."
+
+ _df = pd.DataFrame({"estimate": numeric, "ll": string, "hl": string})
+ with pytest.raises(TypeError) as excinfo:
+ check_data(dataframe=_df, estimate="estimate", varlabel="estimate", ll="ll", hl=None)
+ assert str(excinfo.value) == "'hl' is None. 'll' should also be None."
+
##########################################################################
## Check annote
##########################################################################
diff --git a/tests/test_graph_utils.py b/tests/test_graph_utils.py
index c7cd80c..6065788 100644
--- a/tests/test_graph_utils.py
+++ b/tests/test_graph_utils.py
@@ -230,7 +230,7 @@ def test_format_xticks():
)
# No ticks set
_, ax = plt.subplots()
- ax = format_xticks(input_df, ll="ll", hl="hl", xticks=None, ax=ax)
+ ax = format_xticks(input_df, estimate="estimate", ll="ll", hl="hl", xticks=None, ax=ax)
assert isinstance(ax, Axes)
ax_xmin, ax_xmax = ax.get_xlim()
data_xmin, data_xmax = input_df.ll.min(), input_df.hl.max()
@@ -239,7 +239,9 @@ def test_format_xticks():
# Set xticks
_, ax = plt.subplots()
- ax = format_xticks(input_df, ll="ll", hl="hl", xticks=[1, 2, 3], ax=ax)
+ ax = format_xticks(
+ input_df, estimate="estimate", ll="ll", hl="hl", xticks=[1, 2, 3], ax=ax
+ )
assert isinstance(ax, Axes)
ax_xmin, ax_xmax = ax.get_xlim()
data_xmin, data_xmax = input_df.ll.min(), input_df.hl.max()






