Reconfigurable Inverted Index (Rii): IVFPQ-based fast and memory efficient approximate nearest neighbor search method with a subset-search functionality.
Reference:
- Y. Matsui, R. Hinami, and S. Satoh, "Reconfigurable Inverted Index", ACM Multimedia 2018 (oral). [paper] [project]
 |
 |
|---|---|
| The search can be operated for a subset of a database. | Rii remains fast even after many new items are added. |
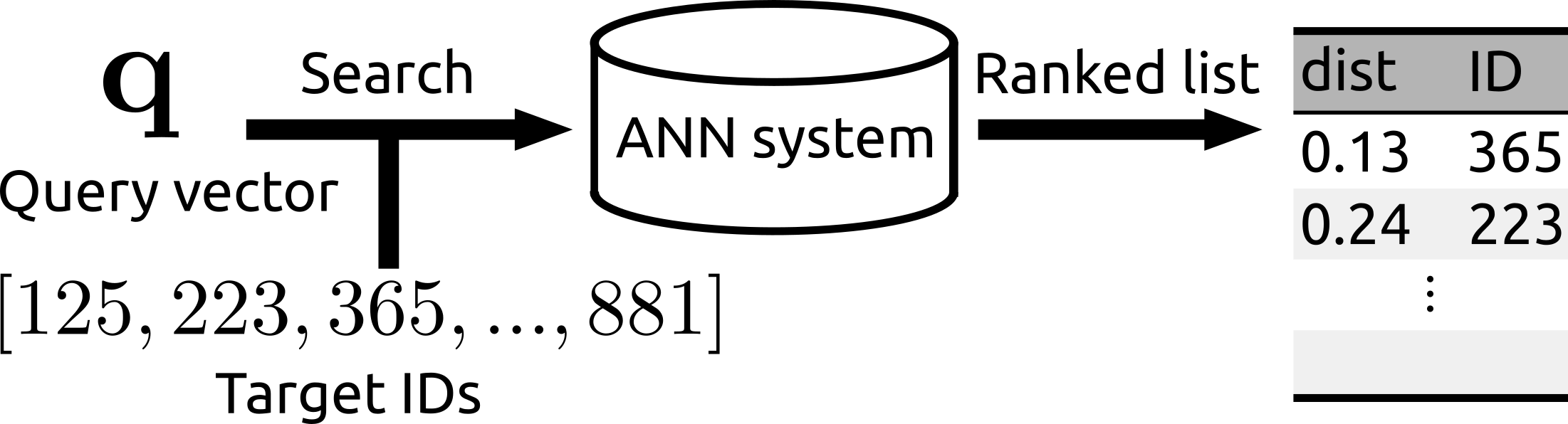
- Fast and memory efficient ANN. Rii enables you to run billion-scale search in less than 10 ms.
- You can run the search over a subset of the whole database
- Rii Remains fast even after many vectors are newly added (i.e., the data structure can be reconfigured)
You can install the package via pip. This library works with Python 3.5+ on linux/mac/wsl/Windows10 (x64, using MSVC:flags - /arch:AVX2, /openmp:llvm, /fp:fast').
pip install rii
For windows (maintained by @ashleyabraham)
pip install https://github.com/ashleyabraham/rii/releases/download/v0.2.7/rii-0.2.7-cp38-cp38-win_amd64.whl
In order to use OpenMP 3.0 /openmp:llvm flag is used which causes warnings of multiple libs loading, use at your descretion when used with other parallel processing library loadings. To supress use
SET KMP_DUPLICATE_LIB_OK=TRUE
The /arch:AVX2 flag is used in MSVC to set appropriate SIMD preprocessors and compiler intrinsics
import rii
import nanopq
import numpy as np
N, Nt, D = 10000, 1000, 128
X = np.random.random((N, D)).astype(np.float32) # 10,000 128-dim vectors to be searched
Xt = np.random.random((Nt, D)).astype(np.float32) # 1,000 128-dim vectors for training
q = np.random.random((D,)).astype(np.float32) # a 128-dim vector
# Prepare a PQ/OPQ codec with M=32 sub spaces
codec = nanopq.PQ(M=32).fit(vecs=Xt) # Trained using Xt
# Instantiate a Rii class with the codec
e = rii.Rii(fine_quantizer=codec)
# Add vectors
e.add_configure(vecs=X)
# Search
ids, dists = e.query(q=q, topk=3)
print(ids, dists) # e.g., [7484 8173 1556] [15.06257439 15.38533878 16.16935158]Note that you can construct a PQ codec and instantiate the Rii class at the same time if you want.
e = rii.Rii(fine_quantizer=nanopq.PQ(M=32).fit(vecs=Xt))
e.add_configure(vecs=X)Furthermore, you can even write them in one line by chaining a function.
e = rii.Rii(fine_quantizer=nanopq.PQ(M=32).fit(vecs=Xt)).add_configure(vecs=X)# The search can be conducted over a subset of the database
target_ids = np.array([85, 132, 236, 551, 694, 728, 992, 1234]) # Specified by IDs
ids, dists = e.query(q=q, topk=3, target_ids=target_ids)
print(ids, dists) # e.g., [728 85 132] [14.80522156 15.92787838 16.28690338]# Add new vectors
X2 = np.random.random((1000, D)).astype(np.float32)
e.add(vecs=X2) # Now N is 11000
e.query(q=q) # Ok. (0.12 msec / query)
# However, if you add quite a lot of vectors, the search might become slower
# because the data structure has been optimized for the initial item size (N=10000)
X3 = np.random.random((1000000, D)).astype(np.float32)
e.add(vecs=X3) # A lot. Now N is 1011000
e.query(q=q) # Slower (0.96 msec/query)
# In such case, run the reconfigure function. That updates the data structure
e.reconfigure()
e.query(q=q) # Ok. (0.21 msec / query)import pickle
with open('rii.pkl', 'wb') as f:
pickle.dump(e, f)
with open('rii.pkl', 'rb') as f:
e_dumped = pickle.load(f) # e_dumped is identical to e# Print the current parameters
e.print_params()
# Delete all PQ-codes and posting lists. fine_quantizer is kept.
e.clear()
# You can switch the verbose flag
e.verbose = False
# You can merge two Rii instances if they have the same fine_quantizer
e1 = rii.Rii(fine_quantizer=codec)
e2 = rii.Rii(fine_quantizer=codec)
e1.add_reconfigure(vecs=X1)
e2.add_reconfigure(vecs=X2)
e1.merge(e2) # Now e1 contains both X1 and X2- The logo is designed by @richardbmx (#4)
- The windows implementation is by @ashleyabraham (#42)
