We're upgrading this package, removing Bower, upgrading dependencies versions, etc, we still need to update this README and docs/ to reflect this. This note will be removed once this is done.
Interactive network visualisation tool for exploration of heterogeneous biological knowledge graphs.
KnetMaps paper at https://f1000research.com/articles/7-1651/v1.
Note:
- KnetMaps feature documentation is available, for new users, in the User_Guide
- How to use KnetMaps in a static web page is explained in the QuickStart_Guide
- A more detailed explanation on how to customize network datasets and stylesheets is available in the Technical_Guide
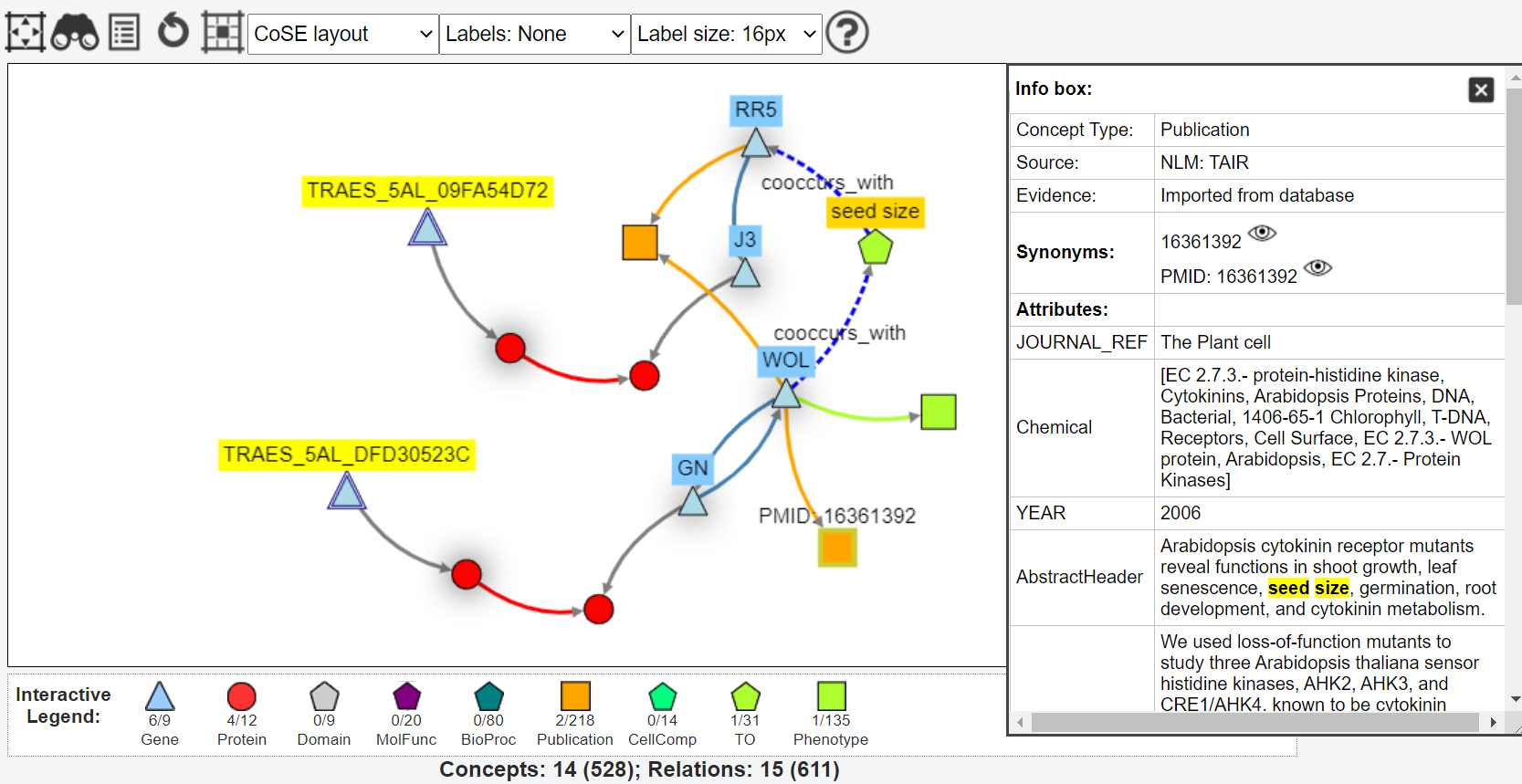
- To explore KnetMaps and its features, check out this Demo example. Instructions to use it are available in the 3 aforementioned KnetMaps guide docs.
- Install Node.js (https://nodejs.org/en/download/) & NPM
- Install Git (https://git-scm.com/)
- Install bower:
npm install -g bower@1.8.8(optional) - Install gulp:
npm install -g gulp@3.9.1 - To build:
gulp optimise - Also, downgrade npm (if errors arise):
npm install -g npm@6.1.0 - Note: Make sure you use
Node v10.21.0as new versions fail with gulp 3 (need gulp 4 which KnetMaps does not support) - Note: Our test environment uses
node 10.21.0, npm 6.1.0 , bower 1.8.8, gulp 3.9.1. Note: As of last check, Node.js 10.7.0 can be downloaded with npm 6.1.0 , which works well with KnetMaps/KnetMiner.
Testing on local PC's is easy and can be done using any lightweight servers like jetty, apache, tomcat, etc.
Python3 users can easily install SimpleHTTPServer via pip and test with that, e.g., on Windows via:
pip install SimpleHTTPServer
cd D:/<path-to-knetmaps-repo>/ #where our `index.html` is located
python -m http.server 9000 #app should be up on http://localhost:9000/index.html