We have developed a structural heterogeneity resolving method for cryo-ET, OPUS-TOMO. Welcome to check out https://github.com/alncat/opusTomo!
This repository contains the implementation of opus-deep structural disentanglement2 (DSD2), which is developed by the research group of Prof. Jianpeng Ma at Fudan University. The preprint of OPUS-DSD2 is available at https://www.biorxiv.org/content/10.1101/2024.09.24.614854v1 and https://drive.google.com/drive/folders/1tEVu9PjCR-4pvkUK17fAHHpyw6y3rZcK?usp=sharing, while the publication of OPUS-DSD is available at https://www.nature.com/articles/s41592-023-02031-6. An exemplar movie of the OPUS-DSD2 is shown below:
mov3.mp4
OPUS-DSD2 also greatly improves the quality of its reconstructions!

The major new functionality of OPUS-DSD2 is reconstructing multi-body dynamics from cryo-EM data end-to-end during structural disentanglement! OPUS-DSD2 can not only disentangle 3D structural information by reconstructing different conformations, but also reconstruct physically meaningful dynamics for the macromolecules. This new function is very easy to use if you have already been familiar with Relion's multi-body refinement (https://elifesciences.org/articles/36861). OPUS-DSD2 takes the input files of Relion's multi-body refinement, then performs structural disentanglement and multi-body dynamics fitting simultaneously and end-to-end!
This program is built upon a set of great works:
This project seeks to unravel how a latent space, encoding 3D structural information, can be learned by utilizing only 2D image supervisions which are aligned against a consensus reference model.
An informative latent space is pivotal as it simplifies data analysis by providing a structured and reduced-dimensional representation of the data.
Our approach strategically leverages the inevitable pose assignment errors introduced during consensus refinement, while concurrently mitigating their impact on the quality of 3D reconstructions. Although it might seem paradoxical to exploit errors while minimizing their effects, our method has proven effective in achieving this delicate balance.
The workflow of OPUS-DSD is demonstrated as follows:

Note that all input and output of this method are in real space! (Fourier space is good, but how about real space!)
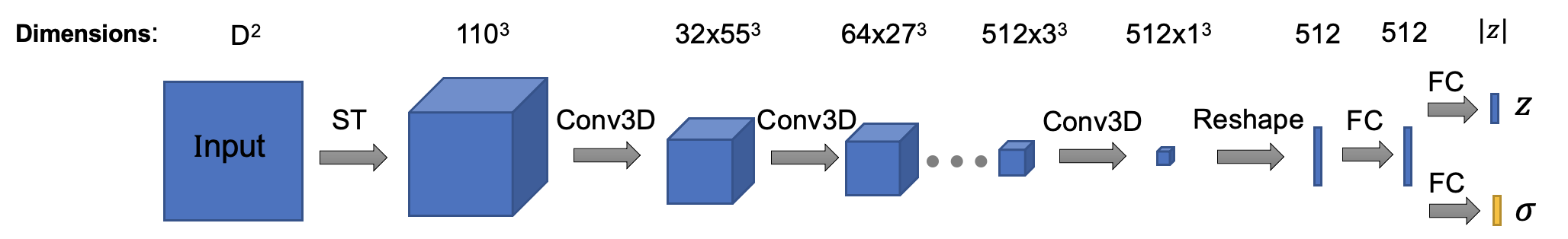
The architecture of encoder is (Encoder class in cryodrgn/models.py):
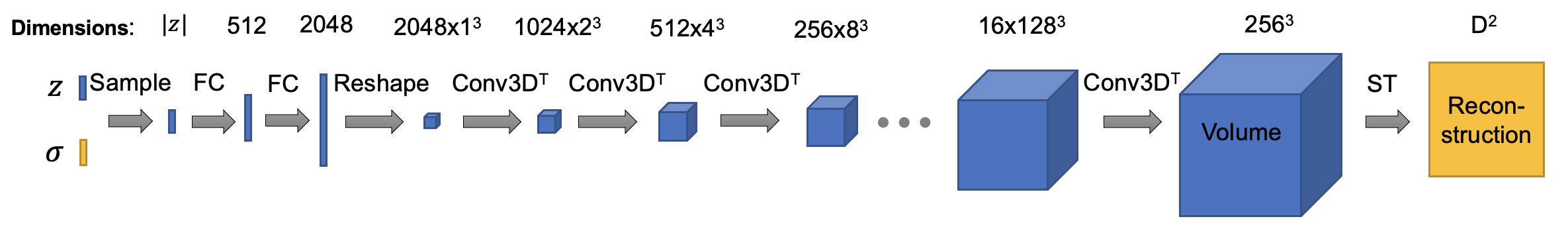
The architecture of composition decoder is (ConvTemplate class in cryodrgn/models.py. In this version, the default size of output volume is set to 192^3, I downsampled the intermediate activations to save some gpu memories. You can tune it as you wish, happy training!):
The architecture of dynamics decoder is:

OPUS-DSD2 has superior structural disentanglement ability to encode distinct compositional changes into different PCs in composition latent space.
mov1.mp4
The open and close of S1 region resolved by OPUS-DSD2 multi-body dynamics, PC3 shows the dynamics to open S1 region, while PC1 shows the dynamics to close S1 region. Red arrows indicate the directions of movements of S1 subunits. Note that these two modes are orthogonal, and have similar patterns of the vibrational dynamics of water molecule H_2O https://strawberryfields.ai/photonics/apps/run_tutorial_dynamics.html
covdefpc3.mp4
The conformational changes inside the NTD of S1 region and S2 region resolved by OPUS-DSD2
covpc3.mp4
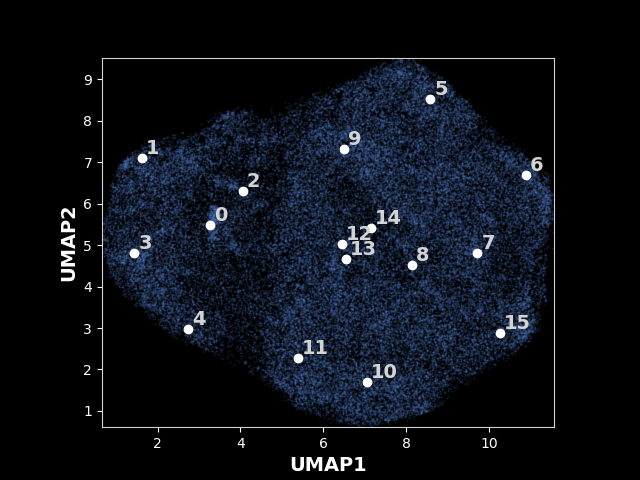
The following results are from the legacy OPUS-DSD. An exmaple UMAP of latent space for 80S ribosome learned by the legacy OPUS-DSD:
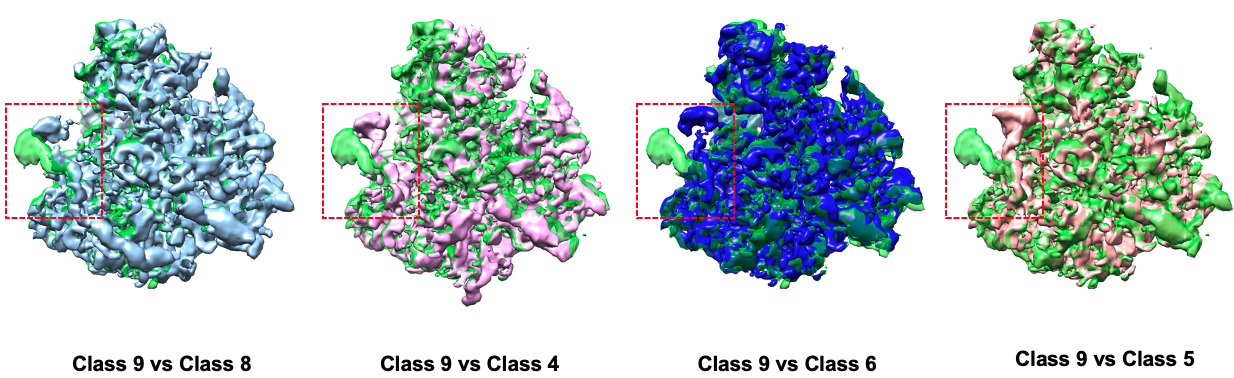
Comparison between some states:
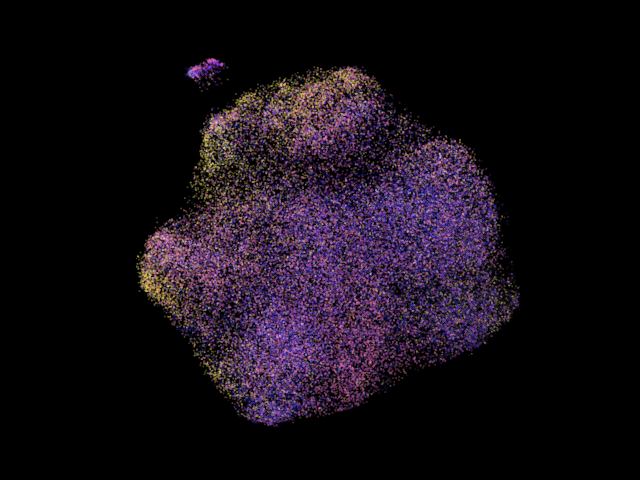
A more colorful umap is shown above. The particles are colored according to their projection classes, note that the clusters often show certain dominant colors, which is due to structural variations in images are accounted in the consensus refinement by distorting the pose paramters of each particle, like fitting a longer rod into a small gap by tilting the rod!
Data source: EMPIAR-10002. The particles are colored according to their pose parameters in this image.
After cloning the repository, to run this program, you need to have an environment with pytorch and a machine with GPUs. The recommended configuration is a machine with 4 V100 GPUs. You can create the conda environment for DSD using one of the environment files in the folder by executing
conda env create --name dsd -f environmentcu11torch11.yml
This environment primarily contains cuda 11.3 and pytorch 1.11.0. To create an environment with cuda 11.3 and pytorch 1.10.1, you can choose environmentcu11.yml. Lastly, environment.yml contains cuda 10.2 and pytorch 1.11.0. On V100 GPU, OPUS-DSD with cuda 11.3 is 20% faster than OPUS-DSD with cuda 10.2. However, it's worth noting that OPUS-DSD has not been tested on Pytorch version higher than 1.11.0. We recommend using pytorch version 1.10.1 or 1.11.0. After the environment is sucessfully created, you can then activate it and execute our program within this environement.
conda activate dsd
You can then install OPUS-DSD by changing to the directory with cloned repository, and execute
pip install -e .
OPUS-DSD can be kept up to date by
git pull
The inference pipeline of our program can run on any GPU which supports cuda 10.2 or 11.3 and is fast to generate a 3D volume. However, the training of our program takes larger amount memory, we recommend using V100 GPUs at least.
This program is developed based on cryoDRGN and adheres to a similar data preparation process.
Data Preparation Guidelines:
- Cryo-EM Dataset: Ensure that the cryo-EM dataset is stored in the MRCS stack file format. Suitable datasets for tutorial are the covid spike protein which is available at: https://pan.baidu.com/s/1PAs7uaUIIeyqegq3sdciYg?pwd=v24j (It contains all required files for training and trained weights), source (https://empiar.pdbj.org/entry/10492)
and the spliceosome which is available at https://empiar.pdbj.org/entry/10180/ (It contains the consensus refinement result.)
- Consensus Refinement Result: The program requires a consensus refinement result, which should not apply any symmetry and must be stored as a Relion STAR file. Other 3D reconstruction results such as 3D classification, as long as they determine the pose parameters of images, can also be supplied as input.
Usage Example:
In overall, the commands for training in OPUS-DSD can be invoked by calling
dsd commandx ...
while the commands for result analysis can be accessed by calling
dsdsh commandx ...
More information about each argument of the command can be displayed using
dsd commandx -h
or
dsdsh commandx -h
Data Preparation for OPUS-DSD Using dsdsh prepare:
There is a command dsdsh prepare for data preparation. Under the hood, dsdsh prepare points to the prepare.sh inside analysis_scripts. Suppose the version of star file is 3.1, the above process can be simplified as,
dsdsh prepare /work/consensus_data.star 320 1.699 --relion31
$1 $2 $3 $4
- $1 specifies the path of the starfile,
- $2 specifies the dimension of image
- $3 specifies the angstrom per pixel of image
- $4 indicates the version of starfile, only include --relion31 if the file version is higher than 3.0
The pose and ctf pkls can be found in the same directory of the starfile, in this case, the pose pkl is /work/consensus_data_pose_euler.pkl, and the ctf pkl is /work/consensus_data_ctf.pkl
Next, you need to prepare a image stack. Suppose you have downloaded the spliceosome dataset. You can prepare a particle stack named all.mrcs using
relion_stack_create --i consensus_data.star --o all --one_by_one
Sometimes after running some protocols in Relion using all.star, Relion might sort the order of images in the corresponding output starfile. You should make sure that the output starfile and the input all.star have the same order of images, thus the output starfile have the correct parameters for the images in all.mrcs!
Finally, you should create a mask using the consensus model and RELION through postprocess. The detailed procedure for mask creation can be found in https://relion.readthedocs.io/en/release-3.1/SPA_tutorial/Mask.html. The spliceosome dataset on empiar comes with a global_mask.mrc file. Suppose the filename of mask is mask.mrc, move it to the program directory for simplicity.
Data Preparation for OPUS-DSD2 with Composition and Dynamics Disentanglement
OPUS-DSD2 features a new capacity to reconstruct multi-body dynamics and resolving compositional heterogeniety. Reconstructing multibody dynamics in OPUS-DSD2 is very similar to Relion's multibody refinement protocol though the underlying dynamics model is different (You can see the details in the preprint). First of all, you shall create a set of masks following Relion's multibody refinement protocol. You can find examples about masks and input starfile for the multibody refinement of spliceosome in https://empiar.pdbj.org/entry/10180/. There is also a tutorial with detailed process for creating masks in https://www.cryst.bbk.ac.uk/embo2019/pracs/RELION%20practical%20EMBO%202019_post%20practice.pdf . The segment map tool in ChimeraX is also perfect for creating segmentations.
After the masks and the starfile for multibody refinement are created, you can prepare the pkls for multibody dynamics estimation by executing
dsdsh prepare_multi starfile D apix masks numb --volumes VOLUMES
The details about each argument can be checked using dsdsh prepare_multi -h
The prepare_multi commands will create a pkl file that contains the parameters of defined bodies, which will be stored in the same directory
as the starfile. The translation of each body is defined using the rotation around its reference body. The magnitude of its translation then is the magnitude of rotation of the center of this body in relative to its reference body. OPUS-DSD2 will read the reference bodies from the starfile, which are specified in _rlnBodyRotateRelativeTo.
But you should note that the index of body starts from 1. The body that occurs most often as the reference body for others will be
selected as the translation-free center. The translation of center will always be set to zero. The direction of rotational axis for the translation of multi-bodies can be determined using
the volume series found by PCA analysis for the OPUS-DSD's result (You can specify the volume series by giving the directory of volume series using --volumes), since they represents a possible mode of movement. Otherwise, the direction of rotational axis will be aligned to the displacement between the center of a body and the center of its center body.
After executing these steps, you have all pkls and files required for running opus-DSD2.
We provided a mask pkl file for covid spike protein in : https://pan.baidu.com/s/1PAs7uaUIIeyqegq3sdciYg?pwd=v24j , which is named as mask_test.pkl.
The segmentation of covid spike protein is shown below:

We provided a mask pkl file for spliceosome using the EMPIAR deposited definiton of multi-bodies in https://drive.google.com/file/d/19fKECY3BDNboXmRUp0fPo9YJf3QarR_u/view?usp=drive_link . In the folder from link https://drive.google.com/drive/folders/1tEVu9PjCR-4pvkUK17fAHHpyw6y3rZcK?usp=sharing, you can also find the pose and ctf pkl and the global mask, which are named as consensus_data_pose_euler.pkl, consensus_data_ctf.pkl and mask.mrc, respectively. We also deposited a trained weight and latent encoding file for visualzation in the same folder. The segmentation of spliceosome given by deposited result is shown below.

Data preparation under the hood
OPUS-DSD follows cryoDRGN's input formats. The pose and ctf parameters for image stack are stored as the python pickle files, aka pkl. Suppose the refinement result is stored as consensus_data.star and the format of the Relion STAR file is below version 3.0,
and the consensus_data.star is located at /work/ directory, you can convert STAR to the pose pkl file by executing the command below:
dsd parse_pose_star /work/consensus_data.star -D 320 --Apix 1.699 -o sp-pose-euler.pkl
where
| argument | explanation |
|---|---|
| -D | the dimension of the particle image in your dataset |
| --Apix | is the angstrom per pixel of you dataset |
| -o | followed by the filename of pose parameter used by our program |
| --relion31 | include this argument if you are using star file from relion with version higher than 3.0 |
Next, you can convert STAR to the ctf pkl file by executing:
dsd parse_ctf_star /work/consensus_data.star -D 320 --Apix 1.699 -o sp-ctf.pkl
For the RELION STAR file with version hgiher than 3.0, you should add --relion31 to the command!
When the inputs are available, you can train the vae for structural disentanglement proposed in OPUS-DSD's paper using
dsd train_cv /work/all.mrcs --ctf ./sp-ctf.pkl --poses ./sp-pose-euler.pkl --lazy-single --pe-type vanilla --encode-mode grad --template-type conv -n 20 -b 12 --zdim 12 --lr 1.e-4 --num-gpus 4 --multigpu --beta-control 2. --beta cos -o /work/sp -r ./mask.mrc --downfrac 0.75 --valfrac 0.25 --lamb 1. --split sp-split.pkl --bfactor 4. --templateres 224
The argument following train_cv specifies the image stack.
The three arguments --pe-type vanilla --encode-mode grad --template-type conv ensure OPUS-DSD is selected! Our program set the default values of those arguments to the values shown in above command.
The functionality of each argument is explained in the table:
| argument | explanation |
|---|---|
| --ctf | ctf parameters of the image stack |
| --poses | pose parameters of the image stack |
| -n | the number of training epoches, each training epoch loops through all images in the training set |
| -b | the number of images for each batch on each gpu, depends on the size of available gpu memory |
| --zdim | the dimension of latent encodings, increase the zdim will improve the fitting capacity of neural network, but may risk of overfitting |
| --lr | the initial learning rate for adam optimizer, 1.e-4 should work fine. |
| --num-gpus | the number of gpus used for training, note that the total number of images in the total batch will be n*num-gpus |
| --multigpu | toggle on the data parallel version |
| --beta-control | the restraint strength of the beta-vae prior, the larger the argument, the stronger the restraint. The scale of beta-control should be propotional to the SNR of dataset. Suitable beta-control might help disentanglement by increasing the magnitude of latent encodings and the sparsity of latent encodings, for more details, check out beta vae paper. In our implementation, we adjust the scale of beta-control automatically based on SNR estimation, possible ranges of this argument are [0.5-4.]. You can use larger beta-control for dataset with higher SNR |
| --beta | the schedule for restraint stengths, cos implements the cyclic annealing schedule and is the default option |
| -o | the directory name for storing results, such as model weights, latent encodings |
| -r | the solvent mask created from consensus model, our program will focus on fitting the contents inside the mask (more specifically, the 2D projection of a 3D mask). Since the majority part of image doesn't contain electron density, using the original image size is wasteful, by specifying a mask, our program will automatically determine a suitable crop rate to keep only the region with densities. |
| --downfrac | the downsampling fraction of input image, the input to network will be downsampled to size of D*downfrac, where D is the original size of image. You can set it according to resolution of consensus model and the templateres you set. |
| --lamb | the restraint strength of structural disentanglement prior proposed in DSD, set it according to the SNR of your dataset, for dataset with high SNR such as ribosome, spliceosome, you can set it to 1. or higher, for dataset with lower SNR, consider lowering it. Possible ranges are [0.1, 3.]. If you find the UMAP of embeedings is exaggeratedly stretched into a ribbon, then the lamb you used during training is too high! |
| --split | the filename for storing the train-validation split of image stack |
| --valfrac | the fraction of images in the validation set, default is 0.1 |
| --bfactor | will apply exp(-bfactor/4 * s^2 * 4*pi^2) decaying to the FT of reconstruction, s is the magnitude of frequency, increase it leads to sharper reconstruction, but takes longer to reveal the part of model with weak density since it actually dampens learning rate, possible ranges are [3, 6]. Consider using higher values for more dynamic structures. We will decay the bfactor slightly in every epoch. This is equivalent to learning rate warming up. |
| --templateres | the size of output volume of our convolutional network, it will be further resampled by spatial transformer before projecting to 2D images. The default value is 192. You may keep it around D*downfrac/0.75, which is larger than the input size. This corresponds to downsampling from the output volume of our network. You can tweak it to other resolutions, larger resolutions can generate sharper density maps, choices are Nx16, where N is integer between 8 and 16 |
| --plot | you can also specify this argument if you want to monitor how the reconstruction progress, our program will display the 2D reconstructions and experimental images after 8 times logging intervals. Namely, you switch to interative mode by including this. The interative mode should be run using command python -m cryodrgn.commands.train_cv |
| --tmp-prefix | the prefix of intermediate reconstructions, default value is tmp. OPUS-DSD will output temporary reconstructions to the root directory of this program when training, whose names are $tmp-prefix.mrc |
| --notinmem | include this arguement to let OPUS-DSD reading image stacks from hard disk during training, this is helpful when a huge dataset cannot fit in the memory |
The plot mode will ouput the following images in the directory where you issued the training command:
Each row shows a selected image and its reconstruction from a batch. In the first row, the first image is the experimental image supplemented to encoder, the second image is a 2D reconstruction blurred by the corresponding CTF, the third image is the correpsonding experimental image after 2D masking.
You can use nohup to let the above command execute in background and use redirections like 1>log 2>err to redirect ouput and error messages to the corresponding files.
Happy Training! Open an issue when running into any troubles.
To restart execution from a checkpoint, you can use
dsd train_cv /work/all.mrcs --ctf ./sp-ctf.pkl --poses ./sp-pose-euler.pkl --lazy-single -n 20 --pe-type vanilla --encode-mode grad --template-type conv -b 12 --zdim 12 --lr 1.e-4 --num-gpus 4 --multigpu --beta-control 2. --beta cos -o /work/sp -r ./mask.mrc --downfrac 0.75 --lamb 1. --valfrac 0.25 --load /work/sp/weights.0.pkl --latents /work/sp/z.0.pkl --split sp-split.pkl --bfactor 4. --templateres 224
| argument | explanation |
|---|---|
| --load | the weight checkpoint from the restarting epoch |
| --latents | the latent encodings from the restarting epoch |
both are in the output directory
During training, opus-DSD will output temporary volumes called tmp*.mrc (or the prefix you specified), you can check the intermediate results by viewing them in Chimera. Opus-DSD uses 3D volume as intermediate representation, so it requires larger amount of memory, v100 gpus will be sufficient for its training. Its training speed is slower, which requires 2 hours on 4 v100 gpus to finish one epoch on dataset with 20k images. By default, opus-DSD reads all images into memory before training, so it may require some more host memories To disable this behavior, you can include --notinmem into the training command.
To reconstruct the multi-body dynamics, you should use the command dsd train_multi, using dsd train_multi -h to check more details. Tho enbale dynamics reconstruction, you shall specify --masks to load the mask pkl with the parameters for each body. An example command is as below:
dsd train_multi /work/all.mrcs --ctf /work/all_ctf.pkl --poses /work/all_pose_euler.pkl -n 20 -b 12 --zdim 12 --lr 1.e-4 --num-gpus 4 --multigpu --beta-control 2. -o ./ -r /work/MaskCreate/job001/mask.mrc --split /work/pkls/sa-split.pkl --lamb 1. --bfactor 3.75 --downfrac 0.75 --valfrac 0.25 --templateres 224 --masks /work/mask_params.pkl --zaffdim 6 --plot
| argument | explanation |
|---|---|
| --zaffdim | controls the dimension of dynamics latent space |
| --masks | the pkl file contains the parameters for bodies of the macromolecule |
If you omit --masks in the train_multi command, OPUS-DSD2 will estimate a global pose correction instead (train_cv also does this).
You can use the analysis scripts in dsdsh to visualize the learned latent space! The analysis procedure is detailed as following.
The analysis scripts can be invoked by calling command like
dsdsh commandx ...
To access detailed usage information for each command, execute the following:
dsdsh commandx -h
The first step is to sample the latent space using kmeans and PCA algorithms. Suppose the training results are in /work/sp,
dsdsh analyze /work/sp 16 4 16
$1 $2 $3 $4
- $1 is the output directory used in training, which stores
weights.*.pkl, z.*.pkl, config.pkl - $2 is the epoch number you would like to analyze,
- $3 is the number of PCs you would like to sample for traversal
- $4 is the number of clusters for kmeans clustering.
The analysis result will be stored in /work/sp/analyze.16, i.e., the output directory plus the epoch number you analyzed, using the above command. You can find the UMAP with the labeled kmeans centers in /work/sp/analyze.16/kmeans16/umap.png and the umap with particles colored by their projection parameter in /work/sp/analyze.16/umap.png .
After executing the above command once, you may skip the lengthy umap embedding laterly by appending --skip-umap to the command in analyze.sh. Our analysis script will read the pickled umap embeddings directly.
The eval_vol command has following options,
If the model is trained by fitting multi-body dynamics, we have a mode, eval_vol, to reconstruct the multi-body dynamics, using the command
dsdsh eval_vol resdir N dpc num apix --masks MASKS --kmeans KMEANS --dfk DFK
This command selects the DFK class from the kmeans{KMEANS} folder as the template volume, which will be deformed according to the dynamics defined by the PC{dpc} of the dynamics latent space. The generated volumes show the dynamics on the selected class along the selected PC, and can be found in defanalyze.{N}/pc{dpc}. Details about each argument can be checked using dsdsh eval_vol -h. As an example, if you execute:
dsdsh eval_vol . 19 dpc 2 2.2 --masks ../mask_test.pkl --kmeans 20 --dfk 4
This will generate volumes along pc 2 in dynamics latent space using the 4th cluster in kmeans20 as template volume.
You can either generate the volume which corresponds to KMeans cluster centroid or traverses the principal component using, (you can check the content of script first, there are two commands, one is used to evaluate volume at kmeans center, another one is for PC traversal, just choose one according to your use case)
dsdsh eval_vol /work/sp 16 kmeans 16 2.2
$1 $2 $3 $4 $5
- $1 is the output directory used in training, which stores
weights.*.pkl, z.*.pkl, config.pkland the clustering result - $2 is the epoch number you just analyzed
- $3 specifies the kind of analysis result where volumes are generated, i.e., kmeans clusters or principal components, use
kmeansfor kmeans clustering, orpcfor principal components - $4 is the number of kmeans clusters (or principal component) you used in analysis
- $5 is the apix of the generated volumes, you can specify a target value
change to directory /work/sp/analyze.16/kmeans16 to checkout the reference*.mrc, which are the reconstructions
correspond to the cluster centroids.
You can use
dsdsh eval_vol /work/sp 16 pc 1 2.2
$1 $2 $3 $4 $5
to generate volumes along pc1. You can check volumes in /work/sp/analyze.16/pc1. You can make a movie using chimerax's vseries feature. An example script for visualizing movie is in analysis_scripts/movie.py. You can show the movie of the volumes by ChimeraX --script "./analysis_scripts/movie.py reference 0.985.
PCs are great for visualizing the main motions and compositional changes of marcomolecules, while KMeans reveals representative conformations in higher qualities.
If you want to invert the handness of reconstruction, you can include --flip to the above commands.
Finally, you can also retrieve the star files for images in each kmeans cluster using
dsdsh parse_pose /work/consensus_data.star 320 1.699 /work/sp 16 16 --relion31
$1 $2 $3 $4 $5 $6 $7
- $1 is the star file of all images
- $2 is the dimension of image
- $3 is apix value of image
- $4 is the output directory used in training
- $5 is the epoch number you just analyzed
- $6 is the number of kmeans clusters you used in analysis
- $7 indicates the version of starfile, only include this when the version of starfile is higher than 3.0
change to directory /work/sp/analyze.16/kmeans16 to checkout the starfile for images in each cluster.