diff --git a/README.md b/README.md
index 1bd09da808..e6ecfcd8bb 100644
--- a/README.md
+++ b/README.md
@@ -88,6 +88,7 @@ Within the following table, we summarized the current NNI capabilities, we are g
Auto-gbdt
Cifar10-pytorch
Scikit-learn
+ EfficientNet
More...
@@ -126,6 +127,7 @@ Within the following table, we summarized the current NNI capabilities, we are g
ENAS
DARTS
P-DARTS
+ CDARTS
Network Morphism
@@ -165,7 +167,7 @@ Within the following table, we summarized the current NNI capabilities, we are g
-
+
@@ -191,18 +193,18 @@ Within the following table, we summarized the current NNI capabilities, we are g
Support TrainingService
Implement TrainingService
-
-
+
+
-## **Install & Verify**
+## **Installation**
-**Install through pip**
+### **Install**
-* We support Linux, MacOS and Windows (local, remote and pai mode) in current stage, Ubuntu 16.04 or higher, MacOS 10.14.1 along with Windows 10.1809 are tested and supported. Simply run the following `pip install` in an environment that has `python >= 3.5`.
+NNI supports and is tested on Ubuntu >= 16.04, macOS >= 10.14.1, and Windows 10 >= 1809. Simply run the following `pip install` in an environment that has `python 64-bit >= 3.5`.
-Linux and MacOS
+Linux or macOS
```bash
python3 -m pip install --upgrade nni
@@ -214,65 +216,39 @@ Windows
python -m pip install --upgrade nni
```
-Note:
-
-* `--user` can be added if you want to install NNI in your home directory, which does not require any special privileges.
-* Currently NNI on Windows support local, remote and pai mode. Anaconda or Miniconda is highly recommended to install NNI on Windows.
-* If there is any error like `Segmentation fault`, please refer to [FAQ](docs/en_US/Tutorial/FAQ.md)
-
-**Install through source code**
-
-* We support Linux (Ubuntu 16.04 or higher), MacOS (10.14.1) and Windows (10.1809) in our current stage.
-
-Linux and MacOS
-
-* Run the following commands in an environment that has `python >= 3.5`, `git` and `wget`.
-
-```bash
- git clone -b v1.3 https://github.com/Microsoft/nni.git
- cd nni
- source install.sh
-```
-
-Windows
-
-* Run the following commands in an environment that has `python >=3.5`, `git` and `PowerShell`
+If you want to try latest code, please [install NNI](docs/en_US/Tutorial/Installation.md) from source code.
-```bash
- git clone -b v1.3 https://github.com/Microsoft/nni.git
- cd nni
- powershell -ExecutionPolicy Bypass -file install.ps1
-```
+For detail system requirements of NNI, please refer to [here](docs/en_US/Tutorial/Installation.md#system-requirements).
-For the system requirements of NNI, please refer to [Install NNI](docs/en_US/Tutorial/Installation.md)
+Note:
-For NNI on Windows, please refer to [NNI on Windows](docs/en_US/Tutorial/NniOnWindows.md)
+* If there is any privilege issue, add `--user` to install NNI in the user directory.
+* Currently NNI on Windows supports local, remote and pai mode. Anaconda or Miniconda is highly recommended to install NNI on Windows.
+* If there is any error like `Segmentation fault`, please refer to [FAQ](docs/en_US/Tutorial/FAQ.md). For FAQ on Windows, please refer to [NNI on Windows](docs/en_US/Tutorial/NniOnWindows.md).
-**Verify install**
+### **Verify installation**
-The following example is an experiment built on TensorFlow. Make sure you have **TensorFlow 1.x installed** before running it. Note that **currently Tensorflow 2.0 is NOT supported**.
+The following example is built on TensorFlow 1.x. Make sure **TensorFlow 1.x is used** when running it.
* Download the examples via clone the source code.
-```bash
- git clone -b v1.3 https://github.com/Microsoft/nni.git
-```
-
-Linux and MacOS
+ ```bash
+ git clone -b v1.3 https://github.com/Microsoft/nni.git
+ ```
* Run the MNIST example.
-```bash
- nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
-```
+ Linux or macOS
-Windows
+ ```bash
+ nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
+ ```
-* Run the MNIST example.
+ Windows
-```bash
- nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
-```
+ ```bash
+ nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
+ ```
* Wait for the message `INFO: Successfully started experiment!` in the command line. This message indicates that your experiment has been successfully started. You can explore the experiment using the `Web UI url`.
@@ -322,9 +298,10 @@ When you submit a pull request, a CLA-bot will automatically determine whether y
This project has adopted the [Microsoft Open Source Code of Conduct](https://opensource.microsoft.com/codeofconduct/). For more information see the Code of [Conduct FAQ](https://opensource.microsoft.com/codeofconduct/faq/) or contact opencode@microsoft.com with any additional questions or comments.
After getting familiar with contribution agreements, you are ready to create your first PR =), follow the NNI developer tutorials to get start:
-* We recommend new contributors to start with ['good first issue'](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen+label%3A%22good+first+issue%22) or ['help-wanted'](https://github.com/microsoft/nni/issues?q=is%3Aopen+is%3Aissue+label%3A%22help+wanted%22), these issues are simple and easy to start.
+* We recommend new contributors to start with simple issues: ['good first issue'](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen+label%3A%22good+first+issue%22) or ['help-wanted'](https://github.com/microsoft/nni/issues?q=is%3Aopen+is%3Aissue+label%3A%22help+wanted%22).
* [NNI developer environment installation tutorial](docs/en_US/Tutorial/SetupNniDeveloperEnvironment.md)
* [How to debug](docs/en_US/Tutorial/HowToDebug.md)
+* If you have any questions on usage, review [FAQ](https://github.com/microsoft/nni/blob/master/docs/en_US/Tutorial/FAQ.md) first, if there are no relevant issues and answers to your question, try contact NNI dev team and users in [Gitter](https://gitter.im/Microsoft/nni?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge) or [File an issue](https://github.com/microsoft/nni/issues/new/choose) on GitHub.
* [Customize your own Tuner](docs/en_US/Tuner/CustomizeTuner.md)
* [Implement customized TrainingService](docs/en_US/TrainingService/HowToImplementTrainingService.md)
* [Implement a new NAS trainer on NNI](https://github.com/microsoft/nni/blob/master/docs/en_US/NAS/NasInterface.md#implement-a-new-nas-trainer-on-nni)
@@ -368,4 +345,3 @@ We encourage researchers and students leverage these projects to accelerate the
## **License**
The entire codebase is under [MIT license](LICENSE)
-
diff --git a/README_zh_CN.md b/README_zh_CN.md
index ec77fcbd50..9aca68dde8 100644
--- a/README_zh_CN.md
+++ b/README_zh_CN.md
@@ -4,7 +4,7 @@
* * *
-[](LICENSE) [](https://msrasrg.visualstudio.com/NNIOpenSource/_build/latest?definitionId=6) [](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen) [](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen+label%3Abug) [](https://github.com/Microsoft/nni/pulls?q=is%3Apr+is%3Aopen) [](https://github.com/Microsoft/nni/releases) [](https://gitter.im/Microsoft/nni?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge) [](https://nni.readthedocs.io/zh/latest/?badge=latest)
+[](LICENSE) [](https://msrasrg.visualstudio.com/NNIOpenSource/_build/latest?definitionId=17&branchName=master) [](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen) [](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen+label%3Abug) [](https://github.com/Microsoft/nni/pulls?q=is%3Apr+is%3Aopen) [](https://github.com/Microsoft/nni/releases) [](https://gitter.im/Microsoft/nni?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge) [](https://nni.readthedocs.io/zh/latest/?badge=latest)
[English](README.md)
@@ -83,6 +83,7 @@ NNI 提供命令行工具以及友好的 WebUI 来管理训练的 Experiment。
Auto-gbdt
Cifar10-pytorch
Scikit-learn
+ EfficientNet
更多...
@@ -121,6 +122,7 @@ NNI 提供命令行工具以及友好的 WebUI 来管理训练的 Experiment。
ENAS
DARTS
P-DARTS
+ CDARTS
Network Morphism
@@ -160,7 +162,7 @@ NNI 提供命令行工具以及友好的 WebUI 来管理训练的 Experiment。
-
+
@@ -186,18 +188,18 @@ NNI 提供命令行工具以及友好的 WebUI 来管理训练的 Experiment。
支持训练平台
实现训练平台
-
-
+
+
-## **安装和验证**
+## **安装**
-**通过 pip 命令安装**
+### **安装**
-* 当前支持 Linux,MacOS 和 Windows(本机,远程,OpenPAI 模式),在 Ubuntu 16.04 或更高版本,MacOS 10.14.1 以及 Windows 10.1809 上进行了测试。 在 `python >= 3.5` 的环境中,只需要运行 `pip install` 即可完成安装。
+NNI 支持并在 Ubuntu >= 16.04, macOS >= 10.14.1, 和 Windows 10 >= 1809 通过了测试。 在 `python 64-bit >= 3.5` 的环境中,只需要运行 `pip install` 即可完成安装。
-Linux 和 macOS
+Linux 或 macOS
```bash
python3 -m pip install --upgrade nni
@@ -209,65 +211,39 @@ Windows
python -m pip install --upgrade nni
```
-注意:
-
-* 如果需要将 NNI 安装到自己的 home 目录中,可使用 `--user`,这样也不需要任何特殊权限。
-* 目前,Windows 上的 NNI 支持本机,远程和 OpenPAI 模式。 强烈推荐使用 Anaconda 或 Miniconda 在 Windows 上安装 NNI。
-* 如果遇到如`Segmentation fault` 这样的任何错误请参考[常见问题](docs/zh_CN/Tutorial/FAQ.md)。
-
-**通过源代码安装**
-
-* 当前支持 Linux(Ubuntu 16.04 或更高版本),MacOS(10.14.1)以及 Windows 10(1809 版)。
-
-Linux 和 MacOS
-
-* 在 `python >= 3.5` 的环境中运行命令: `git` 和 `wget`,确保安装了这两个组件。
-
-```bash
- git clone -b v1.3 https://github.com/Microsoft/nni.git
- cd nni
- source install.sh
-```
-
-Windows
-
-* 在 `python >=3.5` 的环境中运行命令: `git` 和 `PowerShell`,确保安装了这两个组件。
+如果想要尝试最新代码,可通过源代码[安装 NNI](docs/zh_CN/Tutorial/Installation.md)。
-```bash
- git clone -b v1.3 https://github.com/Microsoft/nni.git
- cd nni
- powershell -ExecutionPolicy Bypass -file install.ps1
-```
+有关 NNI 的详细系统要求,参考[这里](docs/zh_CN/Tutorial/Installation.md#system-requirements)。
-参考[安装 NNI](docs/zh_CN/Tutorial/Installation.md) 了解系统需求。
+注意:
-Windows 上参考 [Windows 上使用 NNI](docs/zh_CN/Tutorial/NniOnWindows.md)。
+* 如果遇到任何权限问题,可添加 `--user` 在用户目录中安装 NNI。
+* 目前,Windows 上的 NNI 支持本机,远程和 OpenPAI 模式。 强烈推荐使用 Anaconda 或 Miniconda 在 Windows 上安装 NNI。
+* 如果遇到如 `Segmentation fault` 等错误参考[常见问题](docs/zh_CN/Tutorial/FAQ.md)。 Windows 上的 FAQ 参考[在 Windows 上使用 NNI](docs/zh_CN/Tutorial/NniOnWindows.md)。
-**验证安装**
+### **验证安装**
-以下示例 Experiment 依赖于 TensorFlow 。 在运行前确保安装了 **TensorFlow 1.x**。 注意,**目前不支持 TensorFlow 2.0**。
+以下示例基于 TensorFlow 1.x 。确保运行环境中使用的的是 ** TensorFlow 1.x**。
* 通过克隆源代码下载示例。
-
-```bash
- git clone -b v1.3 https://github.com/Microsoft/nni.git
-```
-
-Linux 和 MacOS
-
-* 运行 MNIST 示例。
-
-```bash
- nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
-```
-
-Windows
+
+ ```bash
+ git clone -b v1.3 https://github.com/Microsoft/nni.git
+ ```
* 运行 MNIST 示例。
-
-```bash
- nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
-```
+
+ Linux 或 macOS
+
+ ```bash
+ nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
+ ```
+
+ Windows
+
+ ```bash
+ nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
+ ```
* 在命令行中等待输出 `INFO: Successfully started experiment!`。 此消息表明 Experiment 已成功启动。 通过命令行输出的 `Web UI url` 来访问 Experiment 的界面。
@@ -319,11 +295,12 @@ You can use these commands to get more information about the experiment
该项目采用了 [ Microsoft 开源行为准则 ](https://opensource.microsoft.com/codeofconduct/)。 有关详细信息,请参阅[常见问题解答](https://opensource.microsoft.com/codeofconduct/faq/),如有任何疑问或意见可联系 opencode@microsoft.com。
-熟悉贡献协议后,即可按照 NNI 开发人员教程,创建第一个 PR =):
+熟悉贡献协议后,即可按照 NNI 开发人员教程,创建第一个 PR:
-* 推荐新贡献者先找到标有 ['good first issue'](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen+label%3A%22good+first+issue%22) 或 ['help-wanted'](https://github.com/microsoft/nni/issues?q=is%3Aopen+is%3Aissue+label%3A%22help+wanted%22) 标签的 Issue。这些都比较简单,可以从这些问题开始。
+* 推荐新贡献者先从简单的问题开始:['good first issue'](https://github.com/Microsoft/nni/issues?q=is%3Aissue+is%3Aopen+label%3A%22good+first+issue%22) 或 ['help-wanted'](https://github.com/microsoft/nni/issues?q=is%3Aopen+is%3Aissue+label%3A%22help+wanted%22)。
* [NNI 开发环境安装教程](docs/zh_CN/Tutorial/SetupNniDeveloperEnvironment.md)
* [如何调试](docs/zh_CN/Tutorial/HowToDebug.md)
+* 如果有使用上的问题,可先查看[常见问题解答](https://github.com/microsoft/nni/blob/master/docs/zh_CN/Tutorial/FAQ.md)。如果没能解决问题,可通过 [Gitter](https://gitter.im/Microsoft/nni?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge&utm_content=badge) 联系 NNI 开发团队或在 GitHub 上 [报告问题](https://github.com/microsoft/nni/issues/new/choose)。
* [自定义 Tuner](docs/zh_CN/Tuner/CustomizeTuner.md)
* [实现定制的训练平台](docs/zh_CN/TrainingService/HowToImplementTrainingService.md)
* [在 NNI 上实现新的 NAS Trainer](https://github.com/microsoft/nni/blob/master/docs/zh_CN/NAS/NasInterface.md#implement-a-new-nas-trainer-on-nni)
@@ -349,7 +326,7 @@ You can use these commands to get more information about the experiment
* [使用 NNI 为 SPTAG 自动调参](docs/zh_CN/CommunitySharings/SptagAutoTune.md)
* [使用 NNI 为 scikit-learn 查找超参](https://towardsdatascience.com/find-thy-hyper-parameters-for-scikit-learn-pipelines-using-microsoft-nni-f1015b1224c1)
* **博客** - [AutoML 工具(Advisor,NNI 与 Google Vizier)的对比](http://gaocegege.com/Blog/%E6%9C%BA%E5%99%A8%E5%AD%A6%E4%B9%A0/katib-new#%E6%80%BB%E7%BB%93%E4%B8%8E%E5%88%86%E6%9E%90) 作者:[@gaocegege](https://github.com/gaocegege) - kubeflow/katib 的设计与实现的总结与分析章节
- * **Blog (中文)** - [NNI 2019 新功能汇总](https://mp.weixin.qq.com/s/7_KRT-rRojQbNuJzkjFMuA) by @squirrelsc
+ * **博客** - [NNI 2019 新功能汇总](https://mp.weixin.qq.com/s/7_KRT-rRojQbNuJzkjFMuA) by @squirrelsc
## **反馈**
diff --git a/docs/en_US/CommunitySharings/NNI_AutoFeatureEng.md b/docs/en_US/CommunitySharings/NNI_AutoFeatureEng.md
new file mode 100644
index 0000000000..40a1e2f8c1
--- /dev/null
+++ b/docs/en_US/CommunitySharings/NNI_AutoFeatureEng.md
@@ -0,0 +1,99 @@
+# NNI review article from Zhihu: - By Garvin Li
+
+The article is by a NNI user on Zhihu forum. In the article, Garvin had shared his experience on using NNI for Automatic Feature Engineering. We think this article is very useful for users who are interested in using NNI for feature engineering. With author's permission, we translated the original article into English.
+
+**原文(source)**: [如何看待微软最新发布的AutoML平台NNI?By Garvin Li](https://www.zhihu.com/question/297982959/answer/964961829?utm_source=wechat_session&utm_medium=social&utm_oi=28812108627968&from=singlemessage&isappinstalled=0)
+
+## 01 Overview of AutoML
+
+In author's opinion, AutoML is not only about hyperparameter optimization, but
+also a process that can target various stages of the machine learning process,
+including feature engineering, NAS, HPO, etc.
+
+## 02 Overview of NNI
+
+NNI (Neural Network Intelligence) is an open source AutoML toolkit from
+Microsoft, to help users design and tune machine learning models, neural network
+architectures, or a complex system’s parameters in an efficient and automatic
+way.
+
+Link:[ https://github.com/Microsoft/nni](https://github.com/Microsoft/nni)
+
+In general, most of Microsoft tools have one prominent characteristic: the
+design is highly reasonable (regardless of the technology innovation degree).
+NNI's AutoFeatureENG basically meets all user requirements of AutoFeatureENG
+with a very reasonable underlying framework design.
+
+## 03 Details of NNI-AutoFeatureENG
+
+>The article is following the github project: [https://github.com/SpongebBob/tabular_automl_NNI](https://github.com/SpongebBob/tabular_automl_NNI).
+
+Each new user could do AutoFeatureENG with NNI easily and efficiently. To exploring the AutoFeatureENG capability, downloads following required files, and then run NNI install through pip.
+
+
+NNI treats AutoFeatureENG as a two-steps-task, feature generation exploration and feature selection. Feature generation exploration is mainly about feature derivation and high-order feature combination.
+
+## 04 Feature Exploration
+
+For feature derivation, NNI offers many operations which could automatically generate new features, which list [as following](https://github.com/SpongebBob/tabular_automl_NNI/blob/master/AutoFEOp.md) :
+
+**count**: Count encoding is based on replacing categories with their counts computed on the train set, also named frequency encoding.
+
+**target**: Target encoding is based on encoding categorical variable values with the mean of target variable per value.
+
+**embedding**: Regard features as sentences, generate vectors using *Word2Vec.*
+
+**crosscout**: Count encoding on more than one-dimension, alike CTR (Click Through Rate).
+
+**aggregete**: Decide the aggregation functions of the features, including min/max/mean/var.
+
+**nunique**: Statistics of the number of unique features.
+
+**histsta**: Statistics of feature buckets, like histogram statistics.
+
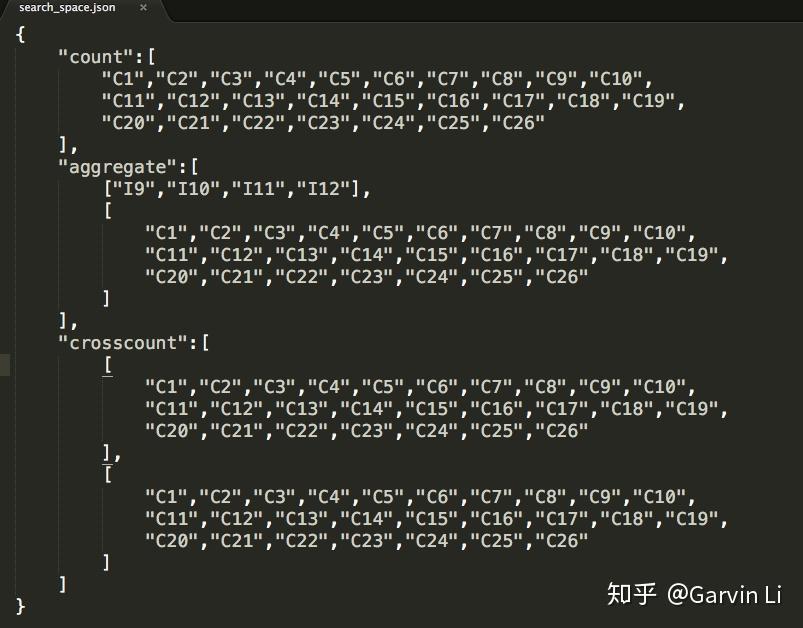
+Search space could be defined in a **JSON file**: to define how specific features intersect, which two columns intersect and how features generate from corresponding columns.
+
+
+
+The picture shows us the procedure of defining search space. NNI provides count encoding for 1-order-op, as well as cross count encoding, aggerate statistics (min max var mean median nunique) for 2-order-op.
+
+For example, we want to search the features which are a frequency encoding (valuecount) features on columns name {“C1”, ...,” C26”}, in the following way:
+
+
+
+we can define a cross frequency encoding (value count on cross dims) method on columns {"C1",...,"C26"} x {"C1",...,"C26"} in the following way:
+
+
+
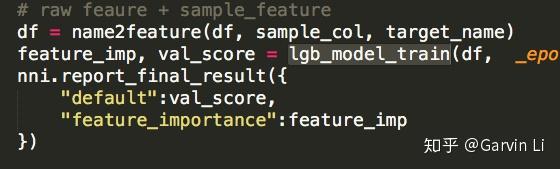
+The purpose of Exploration is to generate new features. You can use **get_next_parameter** function to get received feature candidates of one trial.
+
+>RECEIVED_PARAMS = nni.get_next_parameter()
+
+## 05 Feature selection
+
+To avoid feature explosion and overfitting, feature selection is necessary. In the feature selection of NNI-AutoFeatureENG, LightGBM (Light Gradient Boosting Machine), a gradient boosting framework developed by Microsoft, is mainly promoted.
+
+
+
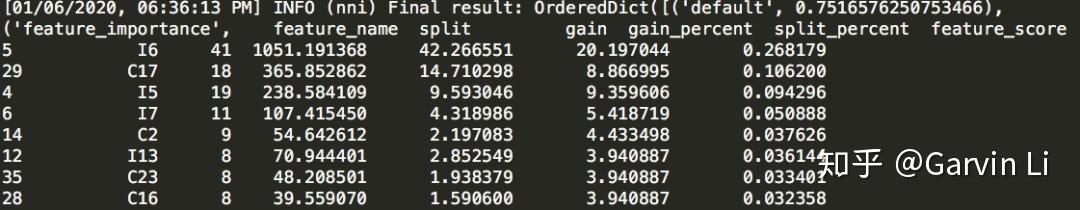
+If you have used **XGBoost** or **GBDT**, you would know the algorithm based on tree structure can easily calculate the importance of each feature on results. LightGBM is able to make feature selection naturally.
+
+The issue is that selected features might be applicable to *GBDT* (Gradient Boosting Decision Tree), but not to the linear algorithm like *LR* (Logistic Regression).
+
+
+
+## 06 Summary
+
+NNI's AutoFeatureEng sets a well-established standard, showing us the operation procedure, available modules, which is highly convenient to use. However, a simple model is probably not enough for good results.
+
+## Suggestions to NNI
+
+About Exploration: If consider using DNN (like xDeepFM) to extract high-order feature would be better.
+
+About Selection: There could be more intelligent options, such as automatic selection system based on downstream models.
+
+Conclusion: NNI could offer users some inspirations of design and it is a good open source project. I suggest researchers leverage it to accelerate the AI research.
+
+Tips: Because the scripts of open source projects are compiled based on gcc7, Mac system may encounter problems of gcc (GNU Compiler Collection). The solution is as follows:
+
+#brew install libomp
+
diff --git a/docs/en_US/CommunitySharings/community_sharings.rst b/docs/en_US/CommunitySharings/community_sharings.rst
index 6938000a9b..23431301c1 100644
--- a/docs/en_US/CommunitySharings/community_sharings.rst
+++ b/docs/en_US/CommunitySharings/community_sharings.rst
@@ -13,3 +13,4 @@ In addtion to the official tutorilas and examples, we encourage community contri
Hyper-parameter Tuning Algorithm Comparsion
Parallelizing Optimization for TPE
Automatically tune systems with NNI
+ NNI review article from Zhihu: - By Garvin Li
diff --git a/docs/en_US/NAS/CDARTS.md b/docs/en_US/NAS/CDARTS.md
new file mode 100644
index 0000000000..4242040f08
--- /dev/null
+++ b/docs/en_US/NAS/CDARTS.md
@@ -0,0 +1,61 @@
+# CDARTS
+
+## Introduction
+
+CDARTS builds a cyclic feedback mechanism between the search and evaluation networks. First, the search network generates an initial topology for evaluation, so that the weights of the evaluation network can be optimized. Second, the architecture topology in the search network is further optimized by the label supervision in classification, as well as the regularization from the evaluation network through feature distillation. Repeating the above cycle results in a joint optimization of the search and evaluation networks, and thus enables the evolution of the topology to fit the final evaluation network.
+
+In implementation of `CdartsTrainer`, it first instantiates two models and two mutators (one for each). The first model is the so-called "search network", which is mutated with a `RegularizedDartsMutator` -- a mutator with subtle differences with `DartsMutator`. The second model is the "evaluation network", which is mutated with a discrete mutator that leverages the previous search network mutator, to sample a single path each time. Trainers train models and mutators alternatively. Users can refer to [references](#reference) if they are interested in more details on these trainers and mutators.
+
+## Reproduction Results
+
+This is CDARTS based on the NNI platform, which currently supports CIFAR10 search and retrain. ImageNet search and retrain should also be supported, and we provide corresponding interfaces. Our reproduced results on NNI are slightly lower than the paper, but much higher than the original DARTS. Here we show the results of three independent experiments on CIFAR10.
+
+| Runs | Paper | NNI |
+| ---- |:-------------:| :-----:|
+| 1 | 97.52 | 97.44 |
+| 2 | 97.53 | 97.48 |
+| 3 | 97.58 | 97.56 |
+
+
+## Examples
+
+[Example code](https://github.com/microsoft/nni/tree/master/examples/nas/cdarts)
+
+```bash
+# In case NNI code is not cloned. If the code is cloned already, ignore this line and enter code folder.
+git clone https://github.com/Microsoft/nni.git
+
+# install apex for distributed training.
+git clone https://github.com/NVIDIA/apex
+cd apex
+python setup.py install --cpp_ext --cuda_ext
+
+# search the best architecture
+cd examples/nas/cdarts
+bash run_search_cifar.sh
+
+# train the best architecture.
+bash run_retrain_cifar.sh
+```
+
+## Reference
+
+### PyTorch
+
+```eval_rst
+.. autoclass:: nni.nas.pytorch.cdarts.CdartsTrainer
+ :members:
+
+ .. automethod:: __init__
+
+.. autoclass:: nni.nas.pytorch.cdarts.RegularizedDartsMutator
+ :members:
+
+.. autoclass:: nni.nas.pytorch.cdarts.DartsDiscreteMutator
+ :members:
+
+ .. automethod:: __init__
+
+.. autoclass:: nni.nas.pytorch.cdarts.RegularizedMutatorParallel
+ :members:
+```
diff --git a/docs/en_US/NAS/Overview.md b/docs/en_US/NAS/Overview.md
index fb3520b5c7..eea44781cc 100644
--- a/docs/en_US/NAS/Overview.md
+++ b/docs/en_US/NAS/Overview.md
@@ -22,6 +22,7 @@ NNI supports below NAS algorithms now and is adding more. User can reproduce an
| [DARTS](DARTS.md) | [DARTS: Differentiable Architecture Search](https://arxiv.org/abs/1806.09055) introduces a novel algorithm for differentiable network architecture search on bilevel optimization. |
| [P-DARTS](PDARTS.md) | [Progressive Differentiable Architecture Search: Bridging the Depth Gap between Search and Evaluation](https://arxiv.org/abs/1904.12760) is based on DARTS. It introduces an efficient algorithm which allows the depth of searched architectures to grow gradually during the training procedure. |
| [SPOS](SPOS.md) | [Single Path One-Shot Neural Architecture Search with Uniform Sampling](https://arxiv.org/abs/1904.00420) constructs a simplified supernet trained with an uniform path sampling method, and applies an evolutionary algorithm to efficiently search for the best-performing architectures. |
+| [CDARTS](CDARTS.md) | [Cyclic Differentiable Architecture Search](https://arxiv.org/abs/****) builds a cyclic feedback mechanism between the search and evaluation networks. It introduces a cyclic differentiable architecture search framework which integrates the two networks into a unified architecture.|
One-shot algorithms run **standalone without nnictl**. Only PyTorch version has been implemented. Tensorflow 2.x will be supported in future release.
diff --git a/docs/en_US/TrainingService/RemoteMachineMode.md b/docs/en_US/TrainingService/RemoteMachineMode.md
index 7e1df06ccc..54a4e45159 100644
--- a/docs/en_US/TrainingService/RemoteMachineMode.md
+++ b/docs/en_US/TrainingService/RemoteMachineMode.md
@@ -1,24 +1,32 @@
-# Run an Experiment on Multiple Machines
+# Run an Experiment on Remote Machines
-NNI supports running an experiment on multiple machines through SSH channel, called `remote` mode. NNI assumes that you have access to those machines, and already setup the environment for running deep learning training code.
+NNI can run one experiment on multiple remote machines through SSH, called `remote` mode. It's like a lightweight training platform. In this mode, NNI can be started from your computer, and dispatch trials to remote machines in parallel.
-e.g. Three machines and you login in with account `bob` (Note: the account is not necessarily the same on different machine):
+## Remote machine requirements
-| IP | Username| Password |
-| -------- |---------|-------|
-| 10.1.1.1 | bob | bob123 |
-| 10.1.1.2 | bob | bob123 |
-| 10.1.1.3 | bob | bob123 |
+* It only supports Linux as remote machines, and [linux part in system specification](../Tutorial/Installation.md) is same as NNI local mode.
-## Setup NNI environment
+* Follow [installation](../Tutorial/Installation.md) to install NNI on each machine.
-Install NNI on each of your machines following the install guide [here](../Tutorial/QuickStart.md).
+* Make sure remote machines meet environment requirements of your trial code. If the default environment does not meet the requirements, the setup script can be added into `command` field of NNI config.
+
+* Make sure remote machines can be accessed through SSH from the machine which runs `nnictl` command. It supports both password and key authentication of SSH. For advanced usages, please refer to [machineList part of configuration](../Tutorial/ExperimentConfig.md).
+
+* Make sure the NNI version on each machine is consistent.
## Run an experiment
-Install NNI on another machine which has network accessibility to those three machines above, or you can just run `nnictl` on any one of the three to launch the experiment.
+e.g. there are three machines, which can be logged in with username and password.
+
+| IP | Username | Password |
+| -------- | -------- | -------- |
+| 10.1.1.1 | bob | bob123 |
+| 10.1.1.2 | bob | bob123 |
+| 10.1.1.3 | bob | bob123 |
+
+Install and run NNI on one of those three machines or another machine, which has network access to them.
-We use `examples/trials/mnist-annotation` as an example here. Shown here is `examples/trials/mnist-annotation/config_remote.yml`:
+Use `examples/trials/mnist-annotation` as the example. Below is content of `examples/trials/mnist-annotation/config_remote.yml`:
```yaml
authorName: default
@@ -58,14 +66,8 @@ machineList:
passwd: bob123
```
-Files in `codeDir` will be automatically uploaded to the remote machine. You can run NNI on different operating systems (Windows, Linux, MacOS) to spawn experiments on the remote machines (only Linux allowed):
+Files in `codeDir` will be uploaded to remote machines automatically. You can run below command on Windows, Linux, or macOS to spawn trials on remote Linux machines:
```bash
nnictl create --config examples/trials/mnist-annotation/config_remote.yml
```
-
-You can also use public/private key pairs instead of username/password for authentication. For advanced usages, please refer to [Experiment Config Reference](../Tutorial/ExperimentConfig.md).
-
-## Version check
-
-NNI support version check feature in since version 0.6, [reference](PaiMode.md).
\ No newline at end of file
diff --git a/docs/en_US/TrainingService/SupportTrainingService.md b/docs/en_US/TrainingService/SupportTrainingService.md
index dfb0df3fe8..ca2b9283fc 100644
--- a/docs/en_US/TrainingService/SupportTrainingService.md
+++ b/docs/en_US/TrainingService/SupportTrainingService.md
@@ -4,10 +4,11 @@ NNI TrainingService provides the training platform for running NNI trial jobs. N
NNI not only provides few built-in training service options, but also provides a method for customers to build their own training service easily.
## Built-in TrainingService
+
|TrainingService|Brief Introduction|
|---|---|
|[__Local__](./LocalMode.md)|NNI supports running an experiment on local machine, called local mode. Local mode means that NNI will run the trial jobs and nniManager process in same machine, and support gpu schedule function for trial jobs.|
-|[__Remote__](./RemoteMachineMode.md)|NNI supports running an experiment on multiple machines through SSH channel, called remote mode. NNI assumes that you have access to those machines, and already setup the environment for running deep learning training code. NNI will submit the trial jobs in remote machine, and schedule suitable machine with enouth gpu resource if specified.|
+|[__Remote__](./RemoteMachineMode.md)|NNI supports running an experiment on multiple machines through SSH channel, called remote mode. NNI assumes that you have access to those machines, and already setup the environment for running deep learning training code. NNI will submit the trial jobs in remote machine, and schedule suitable machine with enough gpu resource if specified.|
|[__Pai__](./PaiMode.md)|NNI supports running an experiment on [OpenPAI](https://github.com/Microsoft/pai) (aka pai), called pai mode. Before starting to use NNI pai mode, you should have an account to access an [OpenPAI](https://github.com/Microsoft/pai) cluster. See [here](https://github.com/Microsoft/pai#how-to-deploy) if you don't have any OpenPAI account and want to deploy an OpenPAI cluster. In pai mode, your trial program will run in pai's container created by Docker.|
|[__Kubeflow__](./KubeflowMode.md)|NNI supports running experiment on [Kubeflow](https://github.com/kubeflow/kubeflow), called kubeflow mode. Before starting to use NNI kubeflow mode, you should have a Kubernetes cluster, either on-premises or [Azure Kubernetes Service(AKS)](https://azure.microsoft.com/en-us/services/kubernetes-service/), a Ubuntu machine on which [kubeconfig](https://kubernetes.io/docs/concepts/configuration/organize-cluster-access-kubeconfig/) is setup to connect to your Kubernetes cluster. If you are not familiar with Kubernetes, [here](https://kubernetes.io/docs/tutorials/kubernetes-basics/) is a good start. In kubeflow mode, your trial program will run as Kubeflow job in Kubernetes cluster.|
|[__FrameworkController__](./FrameworkControllerMode.md)|NNI supports running experiment using [FrameworkController](https://github.com/Microsoft/frameworkcontroller), called frameworkcontroller mode. FrameworkController is built to orchestrate all kinds of applications on Kubernetes, you don't need to install Kubeflow for specific deep learning framework like tf-operator or pytorch-operator. Now you can use FrameworkController as the training service to run NNI experiment.|
@@ -16,7 +17,8 @@ NNI not only provides few built-in training service options, but also provides a
TrainingService is designed to be easily implemented, we define an abstract class TrainingService as the parent class of all kinds of TrainingService, users just need to inherit the parent class and complete their own child class if they want to implement customized TrainingService.
The abstract function in TrainingService is shown below:
-```
+
+```javascript
abstract class TrainingService {
public abstract listTrialJobs(): Promise;
public abstract getTrialJob(trialJobId: string): Promise;
@@ -32,5 +34,6 @@ abstract class TrainingService {
public abstract run(): Promise;
}
```
+
The parent class of TrainingService has a few abstract functions, users need to inherit the parent class and implement all of these abstract functions.
For more information about how to write your own TrainingService, please [refer](https://github.com/microsoft/nni/blob/master/docs/en_US/TrainingService/HowToImplementTrainingService.md).
diff --git a/docs/en_US/TrialExample/EfficientNet.md b/docs/en_US/TrialExample/EfficientNet.md
new file mode 100644
index 0000000000..634a1a9593
--- /dev/null
+++ b/docs/en_US/TrialExample/EfficientNet.md
@@ -0,0 +1,21 @@
+# EfficientNet
+
+[EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks](https://arxiv.org/abs/1905.11946)
+
+Use Grid search to find the best combination of alpha, beta and gamma for EfficientNet-B1, as discussed in Section 3.3 in paper. Search space, tuner, configuration examples are provided here.
+
+## Instructions
+
+[Example code](https://github.com/microsoft/nni/tree/master/examples/trials/efficientnet)
+
+1. Set your working directory here in the example code directory.
+2. Run `git clone https://github.com/ultmaster/EfficientNet-PyTorch` to clone this modified version of [EfficientNet-PyTorch](https://github.com/lukemelas/EfficientNet-PyTorch). The modifications were done to adhere to the original [Tensorflow version](https://github.com/tensorflow/tpu/tree/master/models/official/efficientnet) as close as possible (including EMA, label smoothing and etc.); also added are the part which gets parameters from tuner and reports intermediate/final results. Clone it into `EfficientNet-PyTorch`; the files like `main.py`, `train_imagenet.sh` will appear inside, as specified in the configuration files.
+3. Run `nnictl create --config config_local.yml` (use `config_pai.yml` for OpenPAI) to find the best EfficientNet-B1. Adjust the training service (PAI/local/remote), batch size in the config files according to the environment.
+
+For training on ImageNet, read `EfficientNet-PyTorch/train_imagenet.sh`. Download ImageNet beforehand and extract it adhering to [PyTorch format](https://pytorch.org/docs/stable/torchvision/datasets.html#imagenet) and then replace `/mnt/data/imagenet` in with the location of the ImageNet storage. This file should also be a good example to follow for mounting ImageNet into the container on OpenPAI.
+
+## Results
+
+The follow image is a screenshot, demonstrating the relationship between acc@1 and alpha, beta, gamma.
+
+
diff --git a/docs/en_US/Tuner/HyperbandAdvisor.md b/docs/en_US/Tuner/HyperbandAdvisor.md
index a367b06b13..b7787af199 100644
--- a/docs/en_US/Tuner/HyperbandAdvisor.md
+++ b/docs/en_US/Tuner/HyperbandAdvisor.md
@@ -5,7 +5,7 @@ Hyperband on NNI
[Hyperband][1] is a popular automl algorithm. The basic idea of Hyperband is that it creates several buckets, each bucket has `n` randomly generated hyperparameter configurations, each configuration uses `r` resource (e.g., epoch number, batch number). After the `n` configurations is finished, it chooses top `n/eta` configurations and runs them using increased `r*eta` resource. At last, it chooses the best configuration it has found so far.
## 2. Implementation with fully parallelism
-Frist, this is an example of how to write an automl algorithm based on MsgDispatcherBase, rather than Tuner and Assessor. Hyperband is implemented in this way because it integrates the functions of both Tuner and Assessor, thus, we call it advisor.
+First, this is an example of how to write an automl algorithm based on MsgDispatcherBase, rather than Tuner and Assessor. Hyperband is implemented in this way because it integrates the functions of both Tuner and Assessor, thus, we call it advisor.
Second, this implementation fully leverages Hyperband's internal parallelism. More specifically, the next bucket is not started strictly after the current bucket, instead, it starts when there is available resource.
diff --git a/docs/en_US/Tutorial/FAQ.md b/docs/en_US/Tutorial/FAQ.md
index 6b749f925b..16299e5b20 100644
--- a/docs/en_US/Tutorial/FAQ.md
+++ b/docs/en_US/Tutorial/FAQ.md
@@ -47,5 +47,9 @@ Probably it's a problem with your network config. Here is a checklist.
### NNI on Windows problems
Please refer to [NNI on Windows](NniOnWindows.md)
+
+### More FAQ issues
+[NNI Issues with FAQ labels](https://github.com/microsoft/nni/labels/FAQ)
+
### Help us improve
Please inquiry the problem in https://github.com/Microsoft/nni/issues to see whether there are other people already reported the problem, create a new one if there are no existing issues been created.
diff --git a/docs/en_US/Tutorial/Installation.md b/docs/en_US/Tutorial/Installation.md
index e7711bd2d0..f324366bd8 100644
--- a/docs/en_US/Tutorial/Installation.md
+++ b/docs/en_US/Tutorial/Installation.md
@@ -1,20 +1,22 @@
# Installation of NNI
-Currently we support installation on Linux, Mac and Windows.
+Currently we support installation on Linux, macOS and Windows.
-## **Installation on Linux & Mac**
+## Install on Linux or macOS
-* __Install NNI through pip__
+* Install NNI through pip
- Prerequisite: `python >= 3.5`
+ Prerequisite: `python 64-bit >= 3.5`
```bash
python3 -m pip install --upgrade nni
```
-* __Install NNI through source code__
+* Install NNI through source code
- Prerequisite: `python >=3.5`, `git`, `wget`
+ If you are interested on special or latest code version, you can install NNI through source code.
+
+ Prerequisites: `python 64-bit >=3.5`, `git`, `wget`
```bash
git clone -b v0.8 https://github.com/Microsoft/nni.git
@@ -22,25 +24,27 @@ Currently we support installation on Linux, Mac and Windows.
./install.sh
```
-* __Install NNI in docker image__
+* Use NNI in a docker image
You can also install NNI in a docker image. Please follow the instructions [here](https://github.com/Microsoft/nni/tree/master/deployment/docker/README.md) to build NNI docker image. The NNI docker image can also be retrieved from Docker Hub through the command `docker pull msranni/nni:latest`.
-## **Installation on Windows**
+## Install on Windows
- Anaconda or Miniconda is highly recommended.
+ Anaconda or Miniconda is highly recommended to manage multiple Python environments.
-* __Install NNI through pip__
+* Install NNI through pip
- Prerequisite: `python(64-bit) >= 3.5`
+ Prerequisites: `python 64-bit >= 3.5`
```bash
python -m pip install --upgrade nni
```
-* __Install NNI through source code__
+* Install NNI through source code
+
+ If you are interested on special or latest code version, you can install NNI through source code.
- Prerequisite: `python >=3.5`, `git`, `PowerShell`.
+ Prerequisites: `python 64-bit >=3.5`, `git`, `PowerShell`.
```bash
git clone -b v0.8 https://github.com/Microsoft/nni.git
@@ -48,43 +52,103 @@ Currently we support installation on Linux, Mac and Windows.
powershell -ExecutionPolicy Bypass -file install.ps1
```
-## **System requirements**
-
-Below are the minimum system requirements for NNI on Linux. Due to potential programming changes, the minimum system requirements for NNI may change over time.
-
-||Minimum Requirements|Recommended Specifications|
-|---|---|---|
-|**Operating System**|Ubuntu 16.04 or above|Ubuntu 16.04 or above|
-|**CPU**|Intel® Core™ i3 or AMD Phenom™ X3 8650|Intel® Core™ i5 or AMD Phenom™ II X3 or better|
-|**GPU**|NVIDIA® GeForce® GTX 460|NVIDIA® GeForce® GTX 660 or better|
-|**Memory**|4 GB RAM|6 GB RAM|
-|**Storage**|30 GB available hare drive space|
-|**Internet**|Boardband internet connection|
-|**Resolution**|1024 x 768 minimum display resolution|
-
-Below are the minimum system requirements for NNI on macOS. Due to potential programming changes, the minimum system requirements for NNI may change over time.
-
-||Minimum Requirements|Recommended Specifications|
-|---|---|---|
-|**Operating System**|macOS 10.14.1 (latest version)|macOS 10.14.1 (latest version)|
-|**CPU**|Intel® Core™ i5-760 or better|Intel® Core™ i7-4770 or better|
-|**GPU**|NVIDIA® GeForce® GT 750M or AMD Radeon™ R9 M290 or better|AMD Radeon™ R9 M395X or better|
-|**Memory**|4 GB RAM|8 GB RAM|
-|**Storage**|70GB available space 7200 RPM HDD|70GB available space SSD|
-|**Internet**|Boardband internet connection|
-|**Resolution**|1024 x 768 minimum display resolution|
-
-Below are the minimum system requirements for NNI on Windows, Windows 10.1809 is well tested and recommend. Due to potential programming changes, the minimum system requirements for NNI may change over time.
-
-||Minimum Requirements|Recommended Specifications|
-|---|---|---|
-|**Operating System**|Windows 10|Windows 10|
-|**CPU**|Intel® Core™ i3 or AMD Phenom™ X3 8650|Intel® Core™ i5 or AMD Phenom™ II X3 or better|
-|**GPU**|NVIDIA® GeForce® GTX 460|NVIDIA® GeForce® GTX 660 or better|
-|**Memory**|4 GB RAM|6 GB RAM|
-|**Storage**|30 GB available hare drive space|
-|**Internet**|Boardband internet connection|
-|**Resolution**|1024 x 768 minimum display resolution|
+## Verify installation
+
+The following example is built on TensorFlow 1.x. Make sure **TensorFlow 1.x is used** when running it.
+
+* Download the examples via clone the source code.
+
+ ```bash
+ git clone -b v1.3 https://github.com/Microsoft/nni.git
+ ```
+
+* Run the MNIST example.
+
+ Linux or macOS
+
+ ```bash
+ nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
+ ```
+
+ Windows
+
+ ```bash
+ nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
+ ```
+
+* Wait for the message `INFO: Successfully started experiment!` in the command line. This message indicates that your experiment has been successfully started. You can explore the experiment using the `Web UI url`.
+
+```text
+INFO: Starting restful server...
+INFO: Successfully started Restful server!
+INFO: Setting local config...
+INFO: Successfully set local config!
+INFO: Starting experiment...
+INFO: Successfully started experiment!
+-----------------------------------------------------------------------
+The experiment id is egchD4qy
+The Web UI urls are: http://223.255.255.1:8080 http://127.0.0.1:8080
+-----------------------------------------------------------------------
+
+You can use these commands to get more information about the experiment
+-----------------------------------------------------------------------
+ commands description
+1. nnictl experiment show show the information of experiments
+2. nnictl trial ls list all of trial jobs
+3. nnictl top monitor the status of running experiments
+4. nnictl log stderr show stderr log content
+5. nnictl log stdout show stdout log content
+6. nnictl stop stop an experiment
+7. nnictl trial kill kill a trial job by id
+8. nnictl --help get help information about nnictl
+-----------------------------------------------------------------------
+```
+
+* Open the `Web UI url` in your browser, you can view detail information of the experiment and all the submitted trial jobs as shown below. [Here](../Tutorial/WebUI.md) are more Web UI pages.
+
+
+
+
+
+## System requirements
+
+Due to potential programming changes, the minimum system requirements of NNI may change over time.
+
+### Linux
+
+| | Recommended | Minimum |
+| -------------------- | ---------------------------------------------- | -------------------------------------- |
+| **Operating System** | Ubuntu 16.04 or above |
+| **CPU** | Intel® Core™ i5 or AMD Phenom™ II X3 or better | Intel® Core™ i3 or AMD Phenom™ X3 8650 |
+| **GPU** | NVIDIA® GeForce® GTX 660 or better | NVIDIA® GeForce® GTX 460 |
+| **Memory** | 6 GB RAM | 4 GB RAM |
+| **Storage** | 30 GB available hare drive space |
+| **Internet** | Boardband internet connection |
+| **Resolution** | 1024 x 768 minimum display resolution |
+
+### macOS
+
+| | Recommended | Minimum |
+| -------------------- | ------------------------------------- | --------------------------------------------------------- |
+| **Operating System** | macOS 10.14.1 or above |
+| **CPU** | Intel® Core™ i7-4770 or better | Intel® Core™ i5-760 or better |
+| **GPU** | AMD Radeon™ R9 M395X or better | NVIDIA® GeForce® GT 750M or AMD Radeon™ R9 M290 or better |
+| **Memory** | 8 GB RAM | 4 GB RAM |
+| **Storage** | 70GB available space SSD | 70GB available space 7200 RPM HDD |
+| **Internet** | Boardband internet connection |
+| **Resolution** | 1024 x 768 minimum display resolution |
+
+### Windows
+
+| | Recommended | Minimum |
+| -------------------- | ---------------------------------------------- | -------------------------------------- |
+| **Operating System** | Windows 10 1809 or above |
+| **CPU** | Intel® Core™ i5 or AMD Phenom™ II X3 or better | Intel® Core™ i3 or AMD Phenom™ X3 8650 |
+| **GPU** | NVIDIA® GeForce® GTX 660 or better | NVIDIA® GeForce® GTX 460 |
+| **Memory** | 6 GB RAM | 4 GB RAM |
+| **Storage** | 30 GB available hare drive space |
+| **Internet** | Boardband internet connection |
+| **Resolution** | 1024 x 768 minimum display resolution |
## Further reading
diff --git a/docs/en_US/Tutorial/QuickStart.md b/docs/en_US/Tutorial/QuickStart.md
index f48550cbfc..3e16a46605 100644
--- a/docs/en_US/Tutorial/QuickStart.md
+++ b/docs/en_US/Tutorial/QuickStart.md
@@ -2,14 +2,15 @@
## Installation
-We support Linux MacOS and Windows in current stage, Ubuntu 16.04 or higher, MacOS 10.14.1 and Windows 10.1809 are tested and supported. Simply run the following `pip install` in an environment that has `python >= 3.5`.
-#### Linux and MacOS
+We support Linux macOS and Windows in current stage, Ubuntu 16.04 or higher, macOS 10.14.1 and Windows 10.1809 are tested and supported. Simply run the following `pip install` in an environment that has `python >= 3.5`.
+
+**Linux and macOS**
```bash
python3 -m pip install --upgrade nni
```
-#### Windows
+**Windows**
```bash
python -m pip install --upgrade nni
@@ -17,7 +18,7 @@ We support Linux MacOS and Windows in current stage, Ubuntu 16.04 or higher, Mac
Note:
-* For Linux and MacOS `--user` can be added if you want to install NNI in your home directory, which does not require any special privileges.
+* For Linux and macOS `--user` can be added if you want to install NNI in your home directory, which does not require any special privileges.
* If there is any error like `Segmentation fault`, please refer to [FAQ](FAQ.md)
* For the `system requirements` of NNI, please refer to [Install NNI](Installation.md)
@@ -53,7 +54,7 @@ The above code can only try one set of parameters at a time, if we want to tune
NNI is born for helping user do the tuning jobs, the NNI working process is presented below:
-```
+```text
input: search space, trial code, config file
output: one optimal hyperparameter configuration
@@ -68,7 +69,7 @@ output: one optimal hyperparameter configuration
If you want to use NNI to automatically train your model and find the optimal hyper-parameters, you need to do three changes base on your code:
-**Three things required to do when using NNI**
+**Three steps to start an experiment**
**Step 1**: Give a `Search Space` file in JSON, includes the `name` and the `distribution` (discrete valued or continuous valued) of all the hyperparameters you need to search.
@@ -138,22 +139,25 @@ Note, **for Windows, you need to change trial command `python3` to `python`**
All the codes above are already prepared and stored in [examples/trials/mnist-tfv1/](https://github.com/Microsoft/nni/tree/master/examples/trials/mnist-tfv1).
-#### Linux and MacOS
+**Linux and macOS**
+
Run the **config.yml** file from your command line to start MNIST experiment.
```bash
nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
```
-#### Windows
+
+**Windows**
+
Run the **config_windows.yml** file from your command line to start MNIST experiment.
-**Note**, if you're using NNI on Windows, it needs to change `python3` to `python` in the config.yml file, or use the config_windows.yml file to start the experiment.
+Note, if you're using NNI on Windows, it needs to change `python3` to `python` in the config.yml file, or use the config_windows.yml file to start the experiment.
```bash
nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
```
-Note, **nnictl** is a command line tool, which can be used to control experiments, such as start/stop/resume an experiment, start/stop NNIBoard, etc. Click [here](Nnictl.md) for more usage of `nnictl`
+Note, `nnictl` is a command line tool, which can be used to control experiments, such as start/stop/resume an experiment, start/stop NNIBoard, etc. Click [here](Nnictl.md) for more usage of `nnictl`
Wait for the message `INFO: Successfully started experiment!` in the command line. This message indicates that your experiment has been successfully started. And this is what we expected to get:
@@ -195,7 +199,7 @@ The Web UI urls are: [Your IP]:8080
Open the `Web UI url`(In this information is: `[Your IP]:8080`) in your browser, you can view detail information of the experiment and all the submitted trial jobs as shown below. If you can not open the WebUI link in your terminal, you can refer to [FAQ](FAQ.md).
-#### View summary page
+### View summary page
Click the tab "Overview".
@@ -207,7 +211,7 @@ Top 10 trials will be listed in the Overview page, you can browse all the trials

-#### View trials detail page
+### View trials detail page
Click the tab "Default Metric" to see the point graph of all trials. Hover to see its specific default metric and search space message.
diff --git a/docs/en_US/conf.py b/docs/en_US/conf.py
index 60b2afe782..a8f06f5fc1 100644
--- a/docs/en_US/conf.py
+++ b/docs/en_US/conf.py
@@ -47,6 +47,9 @@
'sphinx.ext.napoleon',
]
+# Add mock modules
+autodoc_mock_imports = ['apex']
+
# Add any paths that contain templates here, relative to this directory.
templates_path = ['_templates']
diff --git a/docs/en_US/examples.rst b/docs/en_US/examples.rst
index d260e648ce..77ea9733b4 100644
--- a/docs/en_US/examples.rst
+++ b/docs/en_US/examples.rst
@@ -12,3 +12,4 @@ Examples
GBDT<./TrialExample/GbdtExample>
RocksDB <./TrialExample/RocksdbExamples>
KDExample <./TrialExample/KDExample>
+ EfficientNet <./TrialExample/EfficientNet>
diff --git a/docs/en_US/nas.rst b/docs/en_US/nas.rst
index 32c235b3bb..a5bd8f6b8f 100644
--- a/docs/en_US/nas.rst
+++ b/docs/en_US/nas.rst
@@ -24,3 +24,4 @@ For details, please refer to the following tutorials:
DARTS
P-DARTS
SPOS
+ CDARTS
diff --git a/examples/trials/efficientnet/assets/search_result.png b/docs/img/efficientnet_search_result.png
similarity index 100%
rename from examples/trials/efficientnet/assets/search_result.png
rename to docs/img/efficientnet_search_result.png
diff --git a/docs/requirements.txt b/docs/requirements.txt
index 35082a1136..54e21d307c 100644
--- a/docs/requirements.txt
+++ b/docs/requirements.txt
@@ -10,4 +10,4 @@ numpy
scipy
coverage
scikit-learn==0.20
-torch==1.3.1
\ No newline at end of file
+https://download.pytorch.org/whl/cpu/torch-1.3.1%2Bcpu-cp37-cp37m-linux_x86_64.whl
diff --git a/docs/zh_CN/CommunitySharings/NNI_AutoFeatureEng.md b/docs/zh_CN/CommunitySharings/NNI_AutoFeatureEng.md
new file mode 100644
index 0000000000..ec932fc3ec
--- /dev/null
+++ b/docs/zh_CN/CommunitySharings/NNI_AutoFeatureEng.md
@@ -0,0 +1,88 @@
+# 来自知乎的评论: - 作者 Garvin Li
+
+本文由 NNI 用户在知乎论坛上发表。 在这篇文章中,Garvin 分享了在使用 NNI 进行自动特征工程方面的体验。 我们认为本文对于有兴趣使用 NNI 进行特征工程的用户非常有用。 经作者许可,将原始文章摘编如下。
+
+**原文**: [如何看待微软最新发布的AutoML平台NNI?作者 Garvin Li](https://www.zhihu.com/question/297982959/answer/964961829?utm_source=wechat_session&utm_medium=social&utm_oi=28812108627968&from=singlemessage&isappinstalled=0)
+
+## 01 AutoML概述
+
+作者认为 AutoML 不光是调参,应该包含自动特征工程。AutoML 是一个系统化的体系,包括:自动特征工程(AutoFeatureEng)、自动调参(AutoTuning)、自动神经网络探索(NAS)等。
+
+## 02 NNI 概述
+
+NNI((Neural Network Intelligence)是一个微软的开源 AutoML 工具包,通过自动而有效的方法来帮助用户设计并调优机器学习模型,神经网络架构,或复杂系统的参数。
+
+链接:[ https://github.com/Microsoft/nni](https://github.com/Microsoft/nni)
+
+我目前只学习了自动特征工程这一个模块,总体看微软的工具都有一个比较大的特点,技术可能不一定多新颖,但是设计都非常赞。 NNI 的 AutoFeatureENG 基本包含了用户对于 AutoFeatureENG 的一切幻想。在微软做 PD 应该挺幸福吧,底层的这些个框架的设计都极为合理。
+
+## 03 细说NNI - AutoFeatureENG
+> 本文使用了此项目: [https://github.com/SpongebBob/tabular_automl_NNI](https://github.com/SpongebBob/tabular_automl_NNI)。
+
+新用户可以使用 NNI 轻松高效地进行 AutoFeatureENG。 使用是非常简单的,安装下文件中的 require,然后 pip install NNI。
+
+ NNI把 AutoFeatureENG 拆分成 exploration 和 selection 两个模块。 exploration 主要是特征衍生和交叉,selection 讲的是如何做特征筛选。
+
+## 04 特征 Exploration
+
+对于功能派生,NNI 提供了许多可自动生成新功能的操作,[列表](https://github.com/SpongebBob/tabular_automl_NNI/blob/master/AutoFEOp.md)如下:
+
+**count**:传统的统计,统计一些数据的出现频率
+
+**target**:特征和目标列的一些映射特征
+
+**embedding**:把特征看成句子,用 *word2vector* 的方式制作向量
+
+**crosscount**:特征间除法,有点类似CTR
+
+**aggregete**:特征的 min/max/var/mean
+
+**nunique**:统计唯一特征的数量。
+
+**histsta**:特征存储桶的统计信息,如直方图统计信息。
+
+具体特征怎么交叉,哪一列和哪一列交叉,每一列特征用什么方式衍生呢?可以通过 **search_space. json** 这个文件控制。
+
+
+
+图片展示了定义搜索空间的过程。 NNI 为 1 阶运算提供计数编码,并为 2 阶运算提供聚合的统计(min max var mean median nunique)。
+
+例如,希望以下列方式搜索列名称 {"C1"、"...","C26"} 上的频率编码(valuecount)功能的功能:
+
+
+
+可以在列 {"C1",...,"C26"} x {"C1",...,"C26"} 上定义交叉频率编码(交叉维度的值计数)方法:
+
+
+
+Exploration 的目的就是长生出新的特征。 在代码里可以用 **get_next_parameter** 的方式获取 tuning 的参数:
+> RECEIVED_PARAMS = nni.get_next_parameter()
+
+## 05 特征 Selection
+
+为了避免特征泛滥的情况,避免过拟合,一定要有 Selection 的机制挑选特征。 在 NNI-AutoFeatureENG 的 Selection 中,主要使用了微软开发的梯度提升框架 LightGBM(Light Gradient Boosting Machine)。
+
+
+
+了解 xgboost 或者 GBDT 算法同学应该知道,这种树形结构的算法是很容易计算出每个特征对于结果的影响的。 所以使用 lightGBM 可以天然的进行特征筛选。
+
+弊病就是,如果下游是个 *LR*(逻辑回归)这种线性算法,筛选出来的特征是否具备普适性。
+
+
+
+## 06 总结
+
+NNI 的 AutoFeature 模块是给整个行业制定了一个教科书般的标准,告诉大家这个东西要怎么做,有哪些模块,使用起来非常方便。 但是如果只是基于这样简单的模式,不一定能达到很好的效果。
+
+## 对 NNI 的建议
+
+我觉得在Exploration方面可以引用一些 DNN(如:xDeepFM) 的特征组合方式,提取更高维度的特征。
+
+在 Selection 方面可以有更多的智能化方案,比如可以基于下游的算法自动选择 Selection 机制。
+
+总之 NNI 在设计曾给了我一些启发,还是一个挺好的开源项目,推荐给大家~ 建议 AI 研究人员使用它来加速研究。
+
+大家用的时候如果是 Mac 电脑可能会遇到 gcc 的问题,因为开源项目自带的脚本是基于 gcc7 编译的, 可以用下面的方法绕过去:
+
+# brew install libomp
+
diff --git a/docs/zh_CN/CommunitySharings/community_sharings.rst b/docs/zh_CN/CommunitySharings/community_sharings.rst
index 828ff48b4d..e549dba143 100644
--- a/docs/zh_CN/CommunitySharings/community_sharings.rst
+++ b/docs/zh_CN/CommunitySharings/community_sharings.rst
@@ -13,3 +13,4 @@
超参调优算法的对比
TPE 的并行优化
使用 NNI 自动调优系统
+ 来自知乎的评论:作者 Garvin Li
diff --git a/docs/zh_CN/Compressor/Pruner.md b/docs/zh_CN/Compressor/Pruner.md
index 0e7963c9d8..d564109149 100644
--- a/docs/zh_CN/Compressor/Pruner.md
+++ b/docs/zh_CN/Compressor/Pruner.md
@@ -335,5 +335,3 @@ pruner.compress()
- **sparsity:** 卷积过滤器要修剪的百分比。
- **op_types:** 在 ActivationMeanRankFilterPruner 中仅支持 Conv2d。
-
-***
\ No newline at end of file
diff --git a/docs/zh_CN/Compressor/Quantizer.md b/docs/zh_CN/Compressor/Quantizer.md
index d2a571f874..3d63a3b3b9 100644
--- a/docs/zh_CN/Compressor/Quantizer.md
+++ b/docs/zh_CN/Compressor/Quantizer.md
@@ -5,10 +5,9 @@ NNI Compressor 中的 Quantizer
Naive Quantizer 将 Quantizer 权重默认设置为 8 位,可用它来测试量化算法。
### 用法
-tensorflow ```python nni.compression.tensorflow.NaiveQuantizer(model_graph).compress()
-```
pytorch
-```python nni.compression.torch.NaiveQuantizer(model).compress()
+```python
+model = nni.compression.torch.NaiveQuantizer(model).compress()
```
***
@@ -45,7 +44,7 @@ quantizer.compress()
查看示例进一步了解
#### QAT Quantizer 的用户配置
-压缩算法所需的常见配置可在[通用配置](./Overview.md#User-configuration-for-a-compression-algorithm)中找到。
+压缩算法所需的常见配置可在[通用配置](./Overview.md#压缩算法中的用户配置)中找到。
此算法所需的配置:
@@ -78,7 +77,7 @@ quantizer.compress()
查看示例进一步了解
#### DoReFa Quantizer 的用户配置
-压缩算法所需的常见配置可在[通用配置](./Overview.md#User-configuration-for-a-compression-algorithm)中找到。
+压缩算法所需的常见配置可在[通用配置](./Overview.md#压缩算法中的用户配置)中找到。
此算法所需的配置:
@@ -114,7 +113,7 @@ model = quantizer.compress()
可以查看示例 [examples/model_compress/BNN_quantizer_cifar10.py](https://github.com/microsoft/nni/tree/master/examples/model_compress/BNN_quantizer_cifar10.py) 了解更多信息。
#### BNN Quantizer 的用户配置
-压缩算法所需的常见配置可在[通用配置](./Overview.md#User-configuration-for-a-compression-algorithm)中找到。
+压缩算法所需的常见配置可在[通用配置](./Overview.md#压缩算法中的用户配置)中找到。
此算法所需的配置:
diff --git a/docs/zh_CN/NAS/CDARTS.md b/docs/zh_CN/NAS/CDARTS.md
new file mode 100644
index 0000000000..b4347127e7
--- /dev/null
+++ b/docs/zh_CN/NAS/CDARTS.md
@@ -0,0 +1,61 @@
+# CDARTS
+
+## 介绍
+
+CDARTS 在搜索和评估网络之间构建了循环反馈机制。 首先,搜索网络会生成初始结构用于评估,以便优化评估网络的权重。 然后,通过分类中通过的标签,以及评估网络中特征蒸馏的正则化来进一步优化搜索网络中的架构。 重复上述循环来优化搜索和评估网路,从而使结构得到训练,成为最终的评估网络。
+
+在 `CdartsTrainer` 的实现中,首先分别实例化了两个 Model 和 Mutator。 第一个 Model 被称为"搜索网络",使用 `RegularizedDartsMutator` 来进行变化。它与 `DartsMutator` 稍有差别。 第二个 Model 是“评估网络”,它里用前面搜索网络的 Mutator 来创建了一个离散的 Mutator,来每次采样一条路径。 Trainer 会交替训练 Model 和 Mutator。 如果对 Trainer 和 Mutator 的实现感兴趣,可参考[这里](#reference)。
+
+## 重现结果
+
+这是基于 NNI 平台的 CDARTS,该平台目前支持 CIFAR10 搜索和重新训练。 同时也支持 ImageNet 的搜索和重新训练,并有相应的接口。 在 NNI 上重现的结果略低于论文,但远高于原始 DARTS。 这里展示了在 CIFAR10 上的三个独立实验的结果。
+
+| 运行 | 论文 | NNI |
+| -- |:-----:|:-----:|
+| 1 | 97.52 | 97.44 |
+| 2 | 97.53 | 97.48 |
+| 3 | 97.58 | 97.56 |
+
+
+## 示例
+
+[示例代码](https://github.com/microsoft/nni/tree/master/examples/nas/cdarts)
+
+```bash
+#如果未克隆 NNI 代码。 如果代码已被克隆,请忽略此行并直接进入代码目录。
+git clone https://github.com/Microsoft/nni.git
+
+# 为分布式训练安装 apex
+git clone https://github.com/NVIDIA/apex
+cd apex
+python setup.py install --cpp_ext --cuda_ext
+
+# 搜索最好的架构
+cd examples/nas/cdarts
+bash run_search_cifar.sh
+
+# 训练最好的架构
+bash run_retrain_cifar.sh
+```
+
+## 参考
+
+### PyTorch
+
+```eval_rst
+.. autoclass:: nni.nas.pytorch.cdarts.CdartsTrainer
+ :members:
+
+ .. automethod:: __init__
+
+.. autoclass:: nni.nas.pytorch.cdarts.RegularizedDartsMutator
+ :members:
+
+.. autoclass:: nni.nas.pytorch.cdarts.DartsDiscreteMutator
+ :members:
+
+ .. automethod:: __init__
+
+.. autoclass:: nni.nas.pytorch.cdarts.RegularizedMutatorParallel
+ :members:
+```
diff --git a/docs/zh_CN/NAS/DARTS.md b/docs/zh_CN/NAS/DARTS.md
index 4f350efa9f..c092070dc4 100644
--- a/docs/zh_CN/NAS/DARTS.md
+++ b/docs/zh_CN/NAS/DARTS.md
@@ -1,4 +1,4 @@
-# NNI 中的 DARTS
+# DARTS
## 介绍
@@ -6,13 +6,45 @@
为了实现,作者在小批量中交替优化网络权重和架构权重。 还进一步探讨了使用二阶优化(unroll)来替代一阶,来提高性能的可能性。
-NNI 的实现基于[官方实现](https://github.com/quark0/darts)以及一个[第三方实现](https://github.com/khanrc/pt.darts)。 目前,在 CIFAR10 上从头训练的一阶和二阶优化均已实现。
+NNI 的实现基于[官方实现](https://github.com/quark0/darts)以及一个[第三方实现](https://github.com/khanrc/pt.darts)。 NNI 上的 DARTS 设计为可用于任何搜索空间。 与原始论文一样,为 CIFAR10 实现了 CNN 的搜索空间,来作为 DARTS 的实际示例。
## 重现结果
-为了重现本文的结果,我们做了一阶和二阶优化的实验。 由于时间限制,我们仅从第二阶段重新训练了*一次**最佳架构*。 我们的结果目前与论文的结果相当。 稍后会增加更多结果
+上述示例旨在重现本文中的结果,我们进行了一阶和二阶优化实验。 由于时间限制,我们仅从第二阶段重新训练了*一次**最佳架构*。 我们的结果目前与论文的结果相当。 稍后会增加更多结果
-| | 论文中 | 重现 |
-| ------------ | ------------- | ---- |
-| 一阶 (CIFAR10) | 3.00 +/- 0.14 | 2.78 |
-| 二阶(CIFAR10) | 2.76 +/- 0.09 | 2.89 |
+| | 论文中 | 重现 |
+| ----------- | ------------- | ---- |
+| 一阶(CIFAR10) | 3.00 +/- 0.14 | 2.78 |
+| 二阶(CIFAR10) | 2.76 +/- 0.09 | 2.89 |
+
+## 示例
+
+### CNN 搜索空间
+
+[示例代码](https://github.com/microsoft/nni/tree/master/examples/nas/darts)
+
+```bash
+#如果未克隆 NNI 代码。 如果代码已被克隆,请忽略此行并直接进入代码目录。
+git clone https://github.com/Microsoft/nni.git
+
+# 搜索最好的架构
+cd examples/nas/darts
+python3 search.py
+
+# 训练最好的架构
+python3 retrain.py --arc-checkpoint ./checkpoints/epoch_49.json
+```
+
+## 参考
+
+### PyTorch
+
+```eval_rst
+.. autoclass:: nni.nas.pytorch.darts.DartsTrainer
+ :members:
+
+ .. automethod:: __init__
+
+.. autoclass:: nni.nas.pytorch.darts.DartsMutator
+ :members:
+```
diff --git a/docs/zh_CN/NAS/ENAS.md b/docs/zh_CN/NAS/ENAS.md
index c25b27bc9b..dcfa3ec060 100644
--- a/docs/zh_CN/NAS/ENAS.md
+++ b/docs/zh_CN/NAS/ENAS.md
@@ -1,7 +1,46 @@
-# NNI 中的 ENAS
+# ENAS
## 介绍
论文 [Efficient Neural Architecture Search via Parameter Sharing](https://arxiv.org/abs/1802.03268) 通过在子模型之间共享参数来加速 NAS 过程。 在 ENAS 中,Contoller 学习在大的计算图中搜索最有子图的方式来发现神经网络。 Controller 通过梯度策略训练,从而选择出能在验证集上有最大期望奖励的子图。 同时对与所选子图对应的模型进行训练,以最小化规范交叉熵损失。
-NNI 的实现基于 [Tensorflow 的官方实现](https://github.com/melodyguan/enas),包括了 CIFAR10 上的 Macro/Micro 搜索空间。 NNI 中从头训练的代码还未完成,当前还没有重现结果。
+NNI 基于官方的 [Tensorflow](https://github.com/melodyguan/enas) 实现,包括通用的强化学习的 Controller,以及能交替训练目标网络和 Controller 的 Trainer。 根据论文,也对 CIFAR10 实现了 Macro 和 Micro 搜索空间来展示如何使用 Trainer。 NNI 中从头训练的代码还未完成,当前还没有重现结果。
+
+## 示例
+
+### CIFAR10 Macro/Micro 搜索空间
+
+[示例代码](https://github.com/microsoft/nni/tree/master/examples/nas/enas)
+
+```bash
+#如果未克隆 NNI 代码。 如果代码已被克隆,请忽略此行并直接进入代码目录。
+git clone https://github.com/Microsoft/nni.git
+
+# 搜索最好的网络架构
+cd examples/nas/enas
+
+# 在 Macro 搜索空间中搜索
+python3 search.py --search-for macro
+
+# 在 Micro 搜索空间中搜索
+python3 search.py --search-for micro
+
+# 查看更多选项
+python3 search.py -h
+```
+
+## 参考
+
+### PyTorch
+
+```eval_rst
+.. autoclass:: nni.nas.pytorch.enas.EnasTrainer
+ :members:
+
+ .. automethod:: __init__
+
+.. autoclass:: nni.nas.pytorch.enas.EnasMutator
+ :members:
+
+ .. automethod:: __init__
+```
diff --git a/docs/zh_CN/NAS/NasInterface.md b/docs/zh_CN/NAS/NasInterface.md
index c7893036d9..dd3f98499f 100644
--- a/docs/zh_CN/NAS/NasInterface.md
+++ b/docs/zh_CN/NAS/NasInterface.md
@@ -98,7 +98,7 @@ trainer.export(file='./chosen_arch')
不同的 Trainer 可能有不同的输入参数,具体取决于其算法。 详细参数可参考具体的 [Trainer 代码](https://github.com/microsoft/nni/tree/master/src/sdk/pynni/nni/nas/pytorch)。 训练完成后,可通过 `trainer.export()` 导出找到的最好的模型。 无需通过 `nnictl` 来启动 NNI Experiment。
-[这里](Overview.md#supported-one-shot-nas-algorithms)是所有支持的 Trainer。 [这里](https://github.com/microsoft/nni/tree/master/examples/nas/simple/train.py)是使用 NNI NAS API 的简单示例。
+[这里](Overview.md#支持的-one-shot-nas-算法)是所有支持的 Trainer。 [这里](https://github.com/microsoft/nni/tree/master/examples/nas/simple/train.py)是使用 NNI NAS API 的简单示例。
### 经典分布式搜索
diff --git a/docs/zh_CN/NAS/Overview.md b/docs/zh_CN/NAS/Overview.md
index 1474a4d788..fc6c734c81 100644
--- a/docs/zh_CN/NAS/Overview.md
+++ b/docs/zh_CN/NAS/Overview.md
@@ -6,93 +6,33 @@
以此为动力,NNI 的目标是提供统一的体系结构,以加速NAS上的创新,并将最新的算法更快地应用于现实世界中的问题上。
-通过[统一的接口](./NasInterface.md),有两种方式进行架构搜索。 [第一种](#supported-one-shot-nas-algorithms)称为 one-shot NAS,基于搜索空间构建了一个超级网络,并使用 one-shot 训练来生成性能良好的子模型。 [第二种](./NasInterface.md#classic-distributed-search)是传统的搜索方法,搜索空间中每个子模型作为独立的 Trial 运行,将性能结果发给 Tuner,由 Tuner 来生成新的子模型。
+通过[统一的接口](./NasInterface.md),有两种方式进行架构搜索。 [一种](#supported-one-shot-nas-algorithms)称为 one-shot NAS,基于搜索空间构建了一个超级网络,并使用 one-shot 训练来生成性能良好的子模型。 [第二种](./NasInterface.md#经典分布式搜索)是传统的搜索方法,搜索空间中每个子模型作为独立的 Trial 运行,将性能结果发给 Tuner,由 Tuner 来生成新的子模型。
* [支持的 One-shot NAS 算法](#supported-one-shot-nas-algorithms)
-* [使用 NNI Experiment 的经典分布式 NAS](./NasInterface.md#classic-distributed-search)
+* [使用 NNI Experiment 的经典分布式 NAS](./NasInterface.md#经典分布式搜索)
* [NNI NAS 编程接口](./NasInterface.md)
## 支持的 One-shot NAS 算法
NNI 现在支持以下 NAS 算法,并且正在添加更多算法。 用户可以重现算法或在自己的数据集上使用它。 鼓励用户使用 [NNI API](#use-nni-api) 实现其它算法,以使更多人受益。
-| 名称 | 算法简介 |
-| ------------------- | --------------------------------------------------------------------------------------------------------------------------------------------- |
-| [ENAS](#enas) | Efficient Neural Architecture Search via Parameter Sharing [参考论文](https://arxiv.org/abs/1802.03268) |
-| [DARTS](#darts) | DARTS: Differentiable Architecture Search [参考论文](https://arxiv.org/abs/1806.09055) |
-| [P-DARTS](#p-darts) | Progressive Differentiable Architecture Search: Bridging the Depth Gap between Search and Evaluation [参考论文](https://arxiv.org/abs/1904.12760) |
+| 名称 | 算法简介 |
+| -------------------- | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
+| [ENAS](ENAS.md) | [Efficient Neural Architecture Search via Parameter Sharing](https://arxiv.org/abs/1802.03268). 在 ENAS 中,Contoller 学习在大的计算图中搜索最有子图的方式来发现神经网络。 它通过在子模型间共享参数来实现加速和出色的性能指标。 |
+| [DARTS](DARTS.md) | [DARTS: Differentiable Architecture Search](https://arxiv.org/abs/1806.09055) 引入了一种在两级网络优化中使用的可微分算法。 |
+| [P-DARTS](PDARTS.md) | [Progressive Differentiable Architecture Search: Bridging the Depth Gap between Search and Evaluation](https://arxiv.org/abs/1904.12760) 基于DARTS。 它引入了一种有效的算法,可在搜索过程中逐渐增加搜索的深度。 |
+| [SPOS](SPOS.md) | 论文 [Single Path One-Shot Neural Architecture Search with Uniform Sampling](https://arxiv.org/abs/1904.00420) 构造了一个采用统一的路径采样方法来训练简化的超网络,并使用进化算法来提高搜索神经网络结构的效率。 |
+| [CDARTS](CDARTS.md) | [Cyclic Differentiable Architecture Search](https://arxiv.org/abs/****) 在搜索和评估的网络见构建了循环反馈的机制。 通过引入的循环的可微分架构搜索框架将两个网络集成为一个架构。 |
-注意,这些算法**不需要 nnictl**,独立运行,仅支持 PyTorch。 将来的版本会支持 Tensorflow 2.0。
+One-shot 算法**不需要 nnictl,可单独运行**。 只实现了 PyTorch 版本。 将来的版本会支持 Tensorflow 2.x。
-### 依赖项
+这是运行示例的一些常见依赖项。 PyTorch 需要高于 1.2 才能使用 `BoolTensor`.
* NNI 1.2+
* tensorboard
* PyTorch 1.2+
* git
-### ENAS
-
-[Efficient Neural Architecture Search via Parameter Sharing](https://arxiv.org/abs/1802.03268). 在 ENAS 中,Contoller 学习在大的计算图中搜索最有子图的方式来发现神经网络。 它通过在子模型间共享参数来实现加速和出色的性能指标。
-
-#### 用法
-
-NNI 中的 ENAS 还在开发中,当前仅支持在 CIFAR10 上 Macro/Micro 搜索空间的搜索阶段。 在 PTB 上从头开始训练及其搜索空间尚未完成。 [详细说明](ENAS.md)。
-
-```bash
-#如果未克隆 NNI 代码。 如果代码已被克隆,请忽略此行并直接进入代码目录。
-git clone https://github.com/Microsoft/nni.git
-
-# 搜索最好的网络架构
-cd examples/nas/enas
-
-# 在 Macro 搜索空间中搜索
-python3 search.py --search-for macro
-
-# 在 Micro 搜索空间中搜索
-python3 search.py --search-for micro
-
-# 查看更多选项
-python3 search.py -h
-```
-
-### DARTS
-
-[DARTS: Differentiable Architecture Search](https://arxiv.org/abs/1806.09055) 在算法上的主要贡献是,引入了一种在两级网络优化中使用的可微分算法。 [详细说明](DARTS.md)。
-
-#### 用法
-
-```bash
-#如果未克隆 NNI 代码。 如果代码已被克隆,请忽略此行并直接进入代码目录。
-git clone https://github.com/Microsoft/nni.git
-
-# 搜索最好的架构
-cd examples/nas/darts
-python3 search.py
-
-# 训练最好的架构
-python3 retrain.py --arc-checkpoint ./checkpoints/epoch_49.json
-```
-
-### P-DARTS
-
-[Progressive Differentiable Architecture Search: Bridging the Depth Gap between Search and Evaluation](https://arxiv.org/abs/1904.12760) 基于 [DARTS](#DARTS)。 它在算法上的主要贡献是引入了一种有效的算法,可在搜索过程中逐渐增加搜索的深度。
-
-#### 用法
-
-```bash
-#如果未克隆 NNI 代码。 如果代码已被克隆,请忽略此行并直接进入代码目录。
-git clone https://github.com/Microsoft/nni.git
-
-# 搜索最好的架构
-cd examples/nas/pdarts
-python3 search.py
-
-# 训练最好的架构,过程与 darts 相同。
-cd ../darts
-python3 retrain.py --arc-checkpoint ../pdarts/checkpoints/epoch_2.json
-```
-
## 使用 NNI API
注意,我们正在尝试通过统一的编程接口来支持各种 NAS 算法,当前处于试验阶段。 这意味着当前编程接口将来会有变化。
@@ -104,7 +44,7 @@ python3 retrain.py --arc-checkpoint ../pdarts/checkpoints/epoch_2.json
1. 在设计神经网络时,可能在层、子模型或连接上有多种选择,并且无法确定是其中一种或某些的组合的结果最好。 因此,需要简单的方法来表达候选的层或子模型。
2. 在神经网络上应用 NAS 时,需要统一的方式来表达架构的搜索空间,这样不必为不同的搜索算法来更改代码。
-NNI 提出的 API 在[这里](https://github.com/microsoft/nni/tree/master/src/sdk/pynni/nni/nas/pytorch)。 [这里](https://github.com/microsoft/nni/tree/master/examples/nas/darts)包含了基于此 API 的 NAS 实现示例。
+NNI 提出的 API 在[这里](https://github.com/microsoft/nni/tree/master/src/sdk/pynni/nni/nas/pytorch)。 [这里](https://github.com/microsoft/nni/tree/master/examples/nas/naive)包含了基于此 API 的 NAS 实现示例。
## **参考和反馈**
* 在 GitHub 中[提交此功能的 Bug](https://github.com/microsoft/nni/issues/new?template=bug-report.md);
diff --git a/docs/zh_CN/Release.md b/docs/zh_CN/Release.md
index 8a65ee765e..eba498b2b6 100644
--- a/docs/zh_CN/Release.md
+++ b/docs/zh_CN/Release.md
@@ -1,5 +1,44 @@
# 更改日志
+## 发布 1.3 - 12/30/2019
+
+### 主要功能
+
+#### 支持神经网络架构搜索算法
+
+* [单路径一次性](https://github.com/microsoft/nni/tree/v1.3/examples/nas/spos/)算法和示例
+
+#### 模型压缩算法支持
+
+* [知识蒸馏](https://github.com/microsoft/nni/blob/v1.3/docs/zh_CN/TrialExample/KDExample.md)算法和使用示例
+* Pruners
+ * [L2Filter Pruner](https://github.com/microsoft/nni/blob/master/docs/zh_CN/Compressor/Pruner.md#l2filter-pruner)
+ * [ActivationAPoZRankFilterPruner](https://github.com/microsoft/nni/blob/master/docs/zh_CN/Compressor/Pruner.md#activationapozrankfilterpruner)
+ * [ActivationMeanRankFilterPruner](https://github.com/microsoft/nni/blob/master/docs/zh_CN/Compressor/Pruner.md#activationmeanrankfilterpruner)
+* [BNN Quantizer](https://github.com/microsoft/nni/blob/v1.3/docs/zh_CN/Compressor/Quantizer.md#bnn-quantizer)
+
+#### 训练平台
+
+* OpenPAI 的 NFS 支持
+
+ 从 OpenPAI v0.11开始,HDFS 不再用作默认存储,可将 NFS、AzureBlob 或其他存储用作默认存储。 在本次版本中,NNI 扩展了对 OpenPAI 最近改动的支持,可与 OpenPAI v0.11 及后续版本的默认存储集成。
+
+* Kubeflow 更新适配
+
+ 适配 Kubeflow 0.7 对 tf-operator 的新支持。
+
+### 工程(代码和生成自动化)
+
+* 启用 [ESLint](https://eslint.org/) 静态代码分析。
+
+### 小改动和 Bug 修复
+
+* 正确识别内置 Tuner 和定制 Tuner
+* Dispatcher 基类的日志
+* 修复有时 Tuner、Assessor 的失败会终止 Experiment 的 Bug。
+* 修复本机作为远程计算机的[问题](https://github.com/microsoft/nni/issues/1852)
+* SMAC Tuner 中 Trial 配置的去重 [ticket](https://github.com/microsoft/nni/issues/1364)
+
## 发布 1.2 - 12/02/2019
### 主要功能
@@ -30,7 +69,7 @@
- 文档
- 改进了 NNI API 文档,增加了更多的 docstring。
-### 修复的 Bug
+### Bug 修复
- 修复当失败的 Trial 没有指标时,表格的排序问题。 -Issue #1773
- 页面切换时,保留选择的(最大、最小)状态。 -PR#1710
@@ -42,14 +81,14 @@
### 主要功能
* 新 Tuner: [PPO Tuner](https://github.com/microsoft/nni/blob/v1.1/docs/zh_CN/Tuner/PPOTuner.md)
-* [查看已停止的 Experiment](https://github.com/microsoft/nni/blob/v1.1/docs/zh_CN/Tutorial/Nnictl.md#view)
+* [查看已停止的 Experiment](https://github.com/microsoft/nni/blob/master/docs/zh_CN/Tutorial/Nnictl.md#view)
* Tuner 可使用专门的 GPU 资源(参考[教程](https://github.com/microsoft/nni/blob/v1.1/docs/zh_CN/Tutorial/ExperimentConfig.md)中的 `gpuIndices` 了解详情)
* 改进 WEB 界面
- Trial 详情页面可列出每个 Trial 的超参,以及开始结束时间(需要通过 "add column" 添加)
- 优化大型 Experiment 的显示性能
- 更多示例
- [EfficientNet PyTorch 示例](https://github.com/ultmaster/EfficientNet-PyTorch)
- - [Cifar10 NAS 示例](https://github.com/microsoft/nni/blob/v1.1/examples/trials/nas_cifar10/README_zh_CN.md)
+ - [Cifar10 NAS 示例](https://github.com/microsoft/nni/blob/v1.1/examples/trials/nas_cifar10/README.md)
- [模型压缩工具包 - Alpha 发布](https://github.com/microsoft/nni/blob/v1.1/docs/zh_CN/Compressor/Overview.md):我们很高兴的宣布 NNI 的模型压缩工具包发布了。它还处于试验阶段,会根据使用反馈来改进。 诚挚邀请您使用、反馈,或更多贡献
### 修复的 Bug
@@ -62,26 +101,28 @@
### 主要功能
* Tuners 和 Assessors
-
- - 支持自动特征生成和选择 -Issue#877 -PR #1387 + 提供自动特征接口 + 基于 Beam 搜索的 Tuner + [添加 Pakdd 示例](https://github.com/microsoft/nni/tree/master/examples/trials/auto-feature-engineering)
- - 添加并行算法提高 TPE 在高并发下的性能。 -PR #1052
- - 为 hyperband 支持多阶段 -PR #1257
-- 训练平台
-
- - 支持私有 Docker Registry -PR #755
-
- * 改进
- * 增加 RestFUL API 的 Python 包装,支持通过代码获取指标的值 PR #1318
- * 新的 Python API : get_experiment_id(), get_trial_id() -PR #1353 -Issue #1331 & -Issue#1368
- * 优化 NAS 搜索空间 -PR #1393
+ - 支持自动特征生成和选择 -Issue#877 -PR #1387
+ + 提供自动特征接口
+ + 基于 Beam 搜索的 Tuner
+ + [增加 Pakdd 示例](https://github.com/microsoft/nni/tree/master/examples/trials/auto-feature-engineering)
+ + 添加并行算法提高 TPE 在高并发下的性能。 -PR #1052
+ + 为 hyperband 支持多阶段 -PR #1257
++ 训练平台
+
+ - 支持私有 Docker Registry -PR #755
+
+ * 改进
+ * 增加 RestFUL API 的 Python 包装,支持通过代码获取指标的值 PR #1318
+ * 新的 Python API : get_experiment_id(), get_trial_id() -PR #1353 -Issue #1331 & -Issue#1368
+ * 优化 NAS 搜索空间 -PR #1393
+ 使用 _type 统一 NAS 搜索空间 -- "mutable_type"e
+ 更新随机搜索 Tuner
- + 将 gpuNum 设为可选 -Issue #1365
- + 删除 OpenPAI 模式下的 outputDir 和 dataDir 配置 -Issue #1342
- + 在 Kubeflow 模式下创建 Trial 时,codeDir 不再被拷贝到 logDir -Issue #1224
+ + 将 gpuNum 设为可选 -Issue #1365
+ + 删除 OpenPAI 模式下的 outputDir 和 dataDir 配置 -Issue #1342
+ + 在 Kubeflow 模式下创建 Trial 时,codeDir 不再被拷贝到 logDir -Issue #1224
+ Web 门户和用户体验
-
+
- 在 Web 界面的搜索过程中显示最好指标的曲线 -Issue #1218
- 在多阶段 Experiment 中,显示参数列表的当前值 -Issue1210 -PR #1348
- 在 AddColumn 中增加 "Intermediate count" 选项。 -Issue #1210
@@ -90,12 +131,13 @@
- 在命令行中为 nnictl 命令增加详细文档的连接 -Issue #1260
- 用户体验改进:显示 Error 日志 -Issue #1173
- 文档
-
+
- 更新文档结构 -Issue #1231
- - [多阶段文档的改进](AdvancedFeature/MultiPhase.md) -Issue #1233 -PR #1242 + 增加配置示例
- - [Web 界面描述改进](Tutorial/WebUI.md) -PR #1419
+ - [多阶段文档的改进](AdvancedFeature/MultiPhase.md) -Issue #1233 -PR #1242
+ + 添加配置示例
+ + [Web 界面描述改进](Tutorial/WebUI.md) -PR #1419
-### 修复的 Bug
+### Bug 修复
* (Bug 修复)修复 0.9 版本中的链接 -Issue #1236
* (Bug 修复)自动完成脚本
@@ -116,20 +158,22 @@
### 主要功能
-* 通用 NAS 编程接口
+* 生成 NAS 编程接口
* 为 NAS 接口添加 `enas-mode` 和 `oneshot-mode`:[PR #1201](https://github.com/microsoft/nni/pull/1201#issue-291094510)
* [有 Matern 核的高斯 Tuner](Tuner/GPTuner.md)
* 支持多阶段 Experiment
-
+
* 为多阶段 Experiment 增加新的训练平台:pai 模式从 v0.9 开始支持多阶段 Experiment。
- * 为以下内置 Tuner 增加多阶段的功能:
- * TPE, Random Search, Anneal, Naïve Evolution, SMAC, Network Morphism, Metis Tuner。
-
- 有关详细信息,参考[实现多阶段的 Tuner](AdvancedFeature/MultiPhase.md)。
+ * 为以下内置 Tuner 增加多阶段的功能:
+
+
+ * TPE, Random Search, Anneal, Naïve Evolution, SMAC, Network Morphism, Metis Tuner。
+
+ 有关详细信息,参考[实现多阶段的 Tuner](AdvancedFeature/MultiPhase.md)。
* Web 界面
-
+
* 在 Web 界面中可比较 Trial。 有关详细信息,参考[查看 Trial 状态](Tutorial/WebUI.md)
* 允许用户调节 Web 界面的刷新间隔。 有关详细信息,参考[查看概要页面](Tutorial/WebUI.md)
* 更友好的显示中间结果。 有关详细信息,参考[查看 Trial 状态](Tutorial/WebUI.md)
@@ -158,7 +202,7 @@
* 在已经运行非 NNI 任务的 GPU 上也能运行 Trial
* 支持 Kubeflow v1beta2 操作符
* 支持 Kubeflow TFJob/PyTorchJob v1beta2
-* [通用 NAS 编程接口](AdvancedFeature/GeneralNasInterfaces.md)
+* [通用 NAS 编程接口](https://github.com/microsoft/nni/blob/v0.8/docs/zh_CN/GeneralNasInterfaces.md)
* 实现了 NAS 的编程接口,可通过 NNI Annotation 很容易的表达神经网络架构搜索空间
* 提供新命令 `nnictl trial codegen` 来调试 NAS 代码生成部分
* 提供 NAS 编程接口教程,NAS 在 MNIST 上的示例,用于 NAS 的可定制的随机 Tuner
@@ -274,10 +318,10 @@
#### 支持新的 Tuner 和 Assessor
-* 支持新的 [Metis Tuner](Tuner/MetisTuner.md)。 **在线**超参调优的场景下,Metis 算法已经被证明非常有效。
+* 支持新的 [Metis Tuner](Tuner/MetisTuner.md)。 对于**在线**超参调优的场景,Metis 算法已经被证明非常有效。
* 支持 [ENAS customized tuner](https://github.com/countif/enas_nni)。由 GitHub 社区用户所贡献。它是神经网络的搜索算法,能够通过强化学习来学习神经网络架构,比 NAS 的性能更好。
* 支持 [Curve fitting (曲线拟合)Assessor](Assessor/CurvefittingAssessor.md),通过曲线拟合的策略来实现提前终止 Trial。
-* 进一步支持 [Weight Sharing(权重共享)](AdvancedFeature/AdvancedNas.md):为 NAS Tuner 通过 NFS 来提供权重共享。
+* [权重共享的](https://github.com/microsoft/nni/blob/v0.5/docs/AdvancedNAS.md)高级支持:为 NAS Tuner 提供权重共享,当前支持 NFS。
#### 改进训练平台
@@ -361,12 +405,12 @@
### NNICTL 的新功能和更新
* 支持同时运行多个 Experiment。
-
+
在 v0.3 以前,NNI 仅支持一次运行一个 Experiment。 此版本开始,用户可以同时运行多个 Experiment。 每个 Experiment 都需要一个唯一的端口,第一个 Experiment 会像以前版本一样使用默认端口。 需要为其它 Experiment 指定唯一端口:
-
- ```bash
- nnictl create --port 8081 --config
- ```
+
+ ```bash
+ nnictl create --port 8081 --config
+ ```
* 支持更新最大 Trial 的数量。 使用 `nnictl update --help` 了解详情。 或参考 [NNICTL](Tutorial/Nnictl.md) 查看完整帮助。
@@ -375,15 +419,15 @@
* 不兼容的改动:nn.get_parameters() 改为 nni.get_next_parameter。 所有以前版本的示例将无法在 v0.3 上运行,需要重新克隆 NNI 代码库获取新示例。 如果在自己的代码中使用了 NNI,也需要相应的更新。
* 新 API **nni.get_sequence_id()**。 每个 Trial 任务都会被分配一个唯一的序列数字,可通过 nni.get_sequence_id() API 来获取。
-
- ```bash
- git clone -b v0.3 https://github.com/microsoft/nni.git
- ```
+
+ ```bash
+ git clone -b v0.3 https://github.com/microsoft/nni.git
+ ```
* **nni.report_final_result(result)** API 对结果参数支持更多的数据类型。
-
+
可用类型:
-
+
* int
* float
* 包含有 'default' 键值的 dict,'default' 的值必须为 int 或 float。 dict 可以包含任何其它键值对。
@@ -394,11 +438,11 @@
### 新示例
-* 公共的 NNI Docker 映像:
-
- ```bash
- docker pull msranni/nni:latest
- ```
+* 公开的 NNI Docker 映像:
+
+ ```bash
+ docker pull msranni/nni:latest
+ ```
* 新的 Trial 示例:[NNI Sklearn 示例](https://github.com/microsoft/nni/tree/master/examples/trials/sklearn)
diff --git a/docs/zh_CN/TrainingService/PaiYarnMode.md b/docs/zh_CN/TrainingService/PaiYarnMode.md
index c84debfa55..0f930967a2 100644
--- a/docs/zh_CN/TrainingService/PaiYarnMode.md
+++ b/docs/zh_CN/TrainingService/PaiYarnMode.md
@@ -102,7 +102,7 @@ paiYarnConfig:
```
nnictl create --config exp_paiYarn.yml
```
-来在 paiYarn 模式下启动 Experiment。 NNI 会为每个 Trial 创建 OpenPAIYarn 作业,作业名称的格式为 `nni_exp_{experiment_id}_trial_{trial_id}`。 可以在 OpenPAIYarn 集群的网站中看到 NNI 创建的作业,例如: 
+来在 paiYarn 模式下启动 Experiment。 NNI 会为每个 Trial 创建 OpenPAIYarn 作业,作业名称的格式为 `nni_exp_{experiment_id}_trial_{trial_id}`。 可以在 OpenPAIYarn 集群的网站中看到 NNI 创建的作业,例如: 
注意:paiYarn 模式下,NNIManager 会启动 RESTful 服务,监听端口为 NNI 网页服务器的端口加1。 例如,如果网页端口为`8080`,那么 RESTful 服务器会监听在 `8081`端口,来接收运行在 Kubernetes 中的 Trial 作业的指标。 因此,需要在防火墙中启用端口 `8081` 的 TCP 协议,以允许传入流量。
diff --git a/docs/zh_CN/TrainingService/RemoteMachineMode.md b/docs/zh_CN/TrainingService/RemoteMachineMode.md
index eba05921b5..e4b6917f84 100644
--- a/docs/zh_CN/TrainingService/RemoteMachineMode.md
+++ b/docs/zh_CN/TrainingService/RemoteMachineMode.md
@@ -1,8 +1,22 @@
-# 在多机上运行 Experiment
+# 在远程计算机上运行 Experiment
-NNI 支持通过 SSH 通道在多台计算机上运行 Experiment,称为 `remote` 模式。 NNI 需要这些计算机的访问权限,并假定已配置好了深度学习训练环境。
+NNI 可以通过 SSH 在多个远程计算机上运行同一个 Experiment,称为 `remote` 模式。 这就像一个轻量级的训练平台。 在此模式下,可以从计算机启动 NNI,并将 Trial 并行调度到远程计算机。
-例如:有三台服务器,登录账户为 `bob`(注意:账户不必在各台计算机上一致):
+## 远程计算机的要求
+
+* 仅支持 Linux 作为远程计算机,其[配置需求](../Tutorial/Installation.md)与 NNI 本机模式相同。
+
+* 根据[安装文章](../Tutorial/Installation.md),在每台计算机上安装 NNI。
+
+* 确保远程计算机满足 Trial 代码的环境要求。 如果默认环境不符合要求,可以将设置脚本添加到 NNI 配置的 `command` 字段。
+
+* 确保远程计算机能被运行 `nnictl` 命令的计算机通过 SSH 访问。 同时支持 SSH 的密码和密钥验证方法。 有关高级用法,参考[配置](../Tutorial/ExperimentConfig.md)的 machineList 部分。
+
+* 确保每台计算机上的 NNI 版本一致。
+
+## 运行 Experiment
+
+例如,有三台机器,可使用用户名和密码登录。
| IP | 用户名 | 密码 |
| -------- | --- | ------ |
@@ -10,15 +24,9 @@ NNI 支持通过 SSH 通道在多台计算机上运行 Experiment,称为 `remo
| 10.1.1.2 | bob | bob123 |
| 10.1.1.3 | bob | bob123 |
-## 设置 NNI 环境
+在这三台计算机或另一台能访问这些计算机的环境中安装并运行 NNI。
-按照[指南](../Tutorial/QuickStart.md)在每台计算机上安装 NNI。
-
-## 运行 Experiment
-
-将 NNI 安装在可以访问上述三台计算机的网络的另一台计算机上,或者仅在三台计算机中的任何一台上运行 `nnictl` 即可启动 Experiment。
-
-以 `examples/trials/mnist-annotation` 为例。 此处示例在 `examples/trials/mnist-annotation/config_remote.yml`:
+以 `examples/trials/mnist-annotation` 为例。 示例文件 `examples/trials/mnist-annotation/config_remote.yml` 的内容如下:
```yaml
authorName: default
@@ -58,14 +66,8 @@ machineList:
passwd: bob123
```
-`codeDir` 中的文件会被自动上传到远程服务器。 可在不同的操作系统上运行 NNI (Windows, Linux, MacOS),来在远程机器上(仅支持 Linux)运行 Experiment。
+`codeDir` 中的文件会自动上传到远程计算机中。 可在 Windows、Linux 或 macOS 上运行以下命令,在远程 Linux 计算机上启动 Trial:
```bash
nnictl create --config examples/trials/mnist-annotation/config_remote.yml
-```
-
-也可使用公钥/私钥对,而非用户名/密码进行身份验证。 有关高级用法,请参考[实验配置参考](../Tutorial/ExperimentConfig.md)。
-
-## 版本校验
-
-从 0.6 开始,NNI 支持版本校验,详情参考[这里](PaiMode.md)。
\ No newline at end of file
+```
\ No newline at end of file
diff --git a/docs/zh_CN/TrainingService/SupportTrainingService.md b/docs/zh_CN/TrainingService/SupportTrainingService.md
index fbf6f6a2cd..5afcc13020 100644
--- a/docs/zh_CN/TrainingService/SupportTrainingService.md
+++ b/docs/zh_CN/TrainingService/SupportTrainingService.md
@@ -19,21 +19,22 @@ NNI 不仅提供了这些内置的训练平台,还提供了轻松连接自己
TrainingService 在设计上为了便于实现,将平台相关的公共属性抽象成类。用户只需要继承这个抽象类,并根据平台特点实现子类,便能够实现 TrainingService。
TrainingService 的声明如下:
- abstract class TrainingService {
- public abstract listTrialJobs(): Promise;
- public abstract getTrialJob(trialJobId: string): Promise;
- public abstract addTrialJobMetricListener(listener: (metric: TrialJobMetric) => void): void;
- public abstract removeTrialJobMetricListener(listener: (metric: TrialJobMetric) => void): void;
- public abstract submitTrialJob(form: JobApplicationForm): Promise;
- public abstract updateTrialJob(trialJobId: string, form: JobApplicationForm): Promise;
- public abstract get isMultiPhaseJobSupported(): boolean;
- public abstract cancelTrialJob(trialJobId: string, isEarlyStopped?: boolean): Promise;
- public abstract setClusterMetadata(key: string, value: string): Promise;
- public abstract getClusterMetadata(key: string): Promise;
- public abstract cleanUp(): Promise;
- public abstract run(): Promise;
- }
-
+```javascript
+abstract class TrainingService {
+ public abstract listTrialJobs(): Promise;
+ public abstract getTrialJob(trialJobId: string): Promise;
+ public abstract addTrialJobMetricListener(listener: (metric: TrialJobMetric) => void): void;
+ public abstract removeTrialJobMetricListener(listener: (metric: TrialJobMetric) => void): void;
+ public abstract submitTrialJob(form: JobApplicationForm): Promise;
+ public abstract updateTrialJob(trialJobId: string, form: JobApplicationForm): Promise;
+ public abstract get isMultiPhaseJobSupported(): boolean;
+ public abstract cancelTrialJob(trialJobId: string, isEarlyStopped?: boolean): Promise;
+ public abstract setClusterMetadata(key: string, value: string): Promise;
+ public abstract getClusterMetadata(key: string): Promise;
+ public abstract cleanUp(): Promise;
+ public abstract run(): Promise;
+}
+```
TrainingService 的父类有一些抽象函数,用户需要继承父类并实现所有这些抽象函数。
有关如何实现 TrainingService 的更多信息,[参考这里](https://github.com/microsoft/nni/blob/master/docs/zh_CN/TrainingService/HowToImplementTrainingService.md)。
\ No newline at end of file
diff --git a/docs/zh_CN/TrialExample/EfficientNet.md b/docs/zh_CN/TrialExample/EfficientNet.md
new file mode 100644
index 0000000000..bf44c695ab
--- /dev/null
+++ b/docs/zh_CN/TrialExample/EfficientNet.md
@@ -0,0 +1,21 @@
+# EfficientNet
+
+[EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks](https://arxiv.org/abs/1905.11946)
+
+如论文中 3.3 所述,使用遍历搜索来找到 EfficientNet-B1 的 alpha, beta 和 gamma 的最好组合。 搜索空间,Tuner,配置示例如下。
+
+## 说明
+
+[示例代码](https://github.com/microsoft/nni/tree/master/examples/trials/efficientnet)
+
+1. 将示例代码目录设为当前工作目录。
+2. 运行 `git clone https://github.com/ultmaster/EfficientNet-PyTorch` 来克隆修改过的 [EfficientNet-PyTorch](https://github.com/lukemelas/EfficientNet-PyTorch)。 修改尽可能接近原始的 [TensorFlow 版本](https://github.com/tensorflow/tpu/tree/master/models/official/efficientnet) (包括 EMA,标记平滑度等等。);另外添加了代码从 Tuner 获取参数并回调中间和最终结果。 将其 clone 至 `EfficientNet-PyTorch`;`main.py`,`train_imagenet.sh` 等文件会在配置文件中指定的路径。
+3. 运行 `nnictl create --config config_local.yml` (OpenPAI 可使用 `config_pai.yml`) 来找到最好的 EfficientNet-B1。 根据环境来调整训练平台(OpenPAI、本机、远程),batch size。
+
+在 ImageNet 上的训练,可阅读 `EfficientNet-PyTorch/train_imagenet.sh`。 下载 ImageNet,并参考 [PyTorch 格式](https://pytorch.org/docs/stable/torchvision/datasets.html#imagenet) 来解压,然后将 `/mnt/data/imagenet` 替换为 ImageNet 的路径。 此文件也是如何将 ImageNet 挂载到 OpenPAI 容器的示例。
+
+## 结果
+
+下图展示了 acc@1 和 alpha、beta、gamma 之间的关系。
+
+
diff --git a/docs/zh_CN/TrialExample/KDExample.md b/docs/zh_CN/TrialExample/KDExample.md
index 8f669b3d6d..ef91b9b905 100644
--- a/docs/zh_CN/TrialExample/KDExample.md
+++ b/docs/zh_CN/TrialExample/KDExample.md
@@ -30,4 +30,4 @@ for batch_idx, (data, target) in enumerate(train_loader):
* **kd_teacher_model:** 预训练过的教师模型
* **kd_T:** 用于平滑教师模型输出的温度。
-完整代码可在这里找到
\ No newline at end of file
+完整代码[在这里](https://github.com/microsoft/nni/tree/v1.3/examples/model_compress/knowledge_distill/)。
diff --git a/docs/zh_CN/TrialExample/SklearnExamples.md b/docs/zh_CN/TrialExample/SklearnExamples.md
index 36f9b6fa67..e860358040 100644
--- a/docs/zh_CN/TrialExample/SklearnExamples.md
+++ b/docs/zh_CN/TrialExample/SklearnExamples.md
@@ -20,7 +20,7 @@ nnictl create --config ./config.yml
示例使用了数字数据集,它是由 1797 个 8x8 的图片组成,每个图片都是一个手写数字,目标是将图片分为 10 类。
-在这个示例中,使用 SVC 作为模型,并为此模型选择一些参数,包括 `"C", "keral", "degree", "gamma" 和 "coef0"`。 关于这些参数的更多信息,可参考[这里](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html)。
+在这个示例中,使用 SVC 作为模型,并为此模型选择一些参数,包括 `"C", "kernel", "degree", "gamma" 和 "coef0"`。 关于这些参数的更多信息,可参考[这里](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html)。
### 2.2 回归
@@ -63,7 +63,7 @@ nnictl create --config ./config.yml
```json
{
"C": {"_type":"uniform","_value":[0.1, 1]},
- "keral": {"_type":"choice","_value":["linear", "rbf", "poly", "sigmoid"]},
+ "kernel": {"_type":"choice","_value":["linear", "rbf", "poly", "sigmoid"]},
"degree": {"_type":"choice","_value":[1, 2, 3, 4]},
"gamma": {"_type":"uniform","_value":[0.01, 0.1]},
"coef0 ": {"_type":"uniform","_value":[0.01, 0.1]}
@@ -75,7 +75,7 @@ nnictl create --config ./config.yml
```python
params = {
'C': 1.0,
- 'keral': 'linear',
+ 'kernel': 'linear',
'degree': 3,
'gamma': 0.01,
'coef0': 0.01
diff --git a/docs/zh_CN/Tutorial/FAQ.md b/docs/zh_CN/Tutorial/FAQ.md
index bcd7fe7a25..7577248612 100644
--- a/docs/zh_CN/Tutorial/FAQ.md
+++ b/docs/zh_CN/Tutorial/FAQ.md
@@ -56,6 +56,10 @@ nnictl 在执行时,使用 tmp 目录作为临时目录来复制 codeDir 下
参考 [Windows 上使用 NNI](NniOnWindows.md)。
+### 更多常见问题解答
+
+[标有常见问题标签的 Issue](https://github.com/microsoft/nni/labels/FAQ)
+
### 帮助改进
在创建新问题前,请在 https://github.com/Microsoft/nni/issues 查看是否有人已经报告了相似的问题。
\ No newline at end of file
diff --git a/docs/zh_CN/Tutorial/HowToDebug.md b/docs/zh_CN/Tutorial/HowToDebug.md
index 580da25dcd..f2c9c72f8c 100644
--- a/docs/zh_CN/Tutorial/HowToDebug.md
+++ b/docs/zh_CN/Tutorial/HowToDebug.md
@@ -81,4 +81,4 @@ NNI 中有不同的错误类型。 根据严重程度,可分为三类。 当 N
如图,每个 Trial 都有日志路径,可以从中找到 Trial 的日志和 stderr。
-除了 Experiment 级调试之外,NNI 还提供调试单个 Trial 的功能,而无需启动整个 Experiment。 有关调试单个 Trial 代码的更多信息,请参考[独立运行模式](../TrialExample/Trials.md#standalone-mode-for-debug)。
\ No newline at end of file
+除了 Experiment 级调试之外,NNI 还提供调试单个 Trial 的功能,而无需启动整个 Experiment。 有关调试单个 Trial 代码的更多信息,请参考[独立运行模式](../TrialExample/Trials.md#用于调试的独立模式)。
\ No newline at end of file
diff --git a/docs/zh_CN/Tutorial/Installation.md b/docs/zh_CN/Tutorial/Installation.md
index 830676ac1b..9a645cdc86 100644
--- a/docs/zh_CN/Tutorial/Installation.md
+++ b/docs/zh_CN/Tutorial/Installation.md
@@ -1,20 +1,22 @@
# 安装 NNI
-当前支持在 Linux,Mac 和 Windows 下安装。
+当前支持在 Linux,macOS 和 Windows 下安装。
-## **在 Linux 和 Mac 下安装**
+## 在 Linux 或 macOS 上安装
-* **通过 pip 命令安装 NNI**
+* 通过 pip 命令安装 NNI
- 先决条件:`python >= 3.5`
+ 先决条件:`python 64-bit >= 3.5`
```bash
python3 -m pip install --upgrade nni
```
-* **通过源代码安装 NNI**
+* 通过源代码安装 NNI
- 先决条件:`python >=3.5`, `git`, `wget`
+ 如果对某个或最新版本的代码感兴趣,可通过源代码安装 NNI。
+
+ 先决条件:`python 64-bit >=3.5`, `git`, `wget`
```bash
git clone -b v0.8 https://github.com/Microsoft/nni.git
@@ -22,25 +24,27 @@
./install.sh
```
-* **在 docker 映像中安装 NNI**
+* 在 Docker 映像中使用 NNI
也可将 NNI 安装到 docker 映像中。 参考[这里](../deployment/docker/README.md)来生成 NNI 的 Docker 映像。 也可通过此命令从 Docker Hub 中直接拉取 NNI 的映像 `docker pull msranni/nni:latest`。
-## **在 Windows 上安装**
+## 在 Windows 上安装
-推荐使用 Anaconda 或 Miniconda。
+强烈建议使用 Anaconda 或 Miniconda 来管理多个 Python 环境。
-* **通过 pip 命令安装 NNI**
+* 通过 pip 命令安装 NNI
- 先决条件:`python(64-bit) >= 3.5`
+ 先决条件:`python 64-bit >= 3.5`
```bash
python -m pip install --upgrade nni
```
-* **通过源代码安装 NNI**
+* 通过源代码安装 NNI
+
+ 如果对某个或最新版本的代码感兴趣,可通过源代码安装 NNI。
- 先决条件:`python >=3.5`, `git`, `PowerShell`
+ 先决条件:`python 64-bit >=3.5`, `git`, `PowerShell`
```bash
git clone -b v0.8 https://github.com/Microsoft/nni.git
@@ -48,43 +52,104 @@
powershell -ExecutionPolicy Bypass -file install.ps1
```
-## **系统需求**
-
-以下是 NNI 在 Linux 下的最低配置。 由于程序变更,NNI 的最低配置会有所更改。
-
-| | 最低配置 | 推荐配置 |
-| -------- | ------------------------------------- | ----------------------------------------- |
-| **操作系统** | Ubuntu 16.04 或以上版本 | Ubuntu 16.04 或以上版本 |
-| **CPU** | Intel® Core™ i3 或 AMD Phenom™ X3 8650 | Intel® Core™ i5 或 AMD Phenom™ II X3 或更高配置 |
-| **GPU** | NVIDIA® GeForce® GTX 460 | NVIDIA® GeForce® GTX 660 或更高配置 |
-| **内存** | 4 GB | 6 GB |
-| **存储** | 30 GB 可用的磁盘空间 | |
-| **网络** | 宽带连接 | |
-| **分辨率** | 1024 x 768 以上 | |
-
-以下是 NNI 在 MacOS 下的最低配置。 由于程序变更,NNI 的最低配置会有所更改。
-
-| | 最低配置 | 推荐配置 |
-| -------- | -------------------------------------------------- | ------------------------ |
-| **操作系统** | macOS 10.14.1 (最新版本) | macOS 10.14.1 (最新版本) |
-| **CPU** | Intel® Core™ i5-760 或更高 | Intel® Core™ i7-4770 或更高 |
-| **GPU** | NVIDIA® GeForce® GT 750M 或 AMD Radeon™ R9 M290 或更高 | AMD Radeon™ R9 M395X 或更高 |
-| **内存** | 4 GB | 8 GB |
-| **存储** | 70GB 可用空间及 7200 RPM 硬盘 | 70GB 可用空间 SSD 硬盘 |
-| **网络** | 宽带连接 | |
-| **分辨率** | 1024 x 768 以上 | |
-
-以下是 NNI 在 Windows 上的最低配置,推荐使用 Windows 10 1809 版。 由于程序变更,NNI 的最低配置会有所更改。
-
-| | 最低配置 | 推荐配置 |
-| -------- | ------------------------------------- | ----------------------------------------- |
-| **操作系统** | Windows 10 | Windows 10 |
-| **CPU** | Intel® Core™ i3 或 AMD Phenom™ X3 8650 | Intel® Core™ i5 或 AMD Phenom™ II X3 或更高配置 |
-| **GPU** | NVIDIA® GeForce® GTX 460 | NVIDIA® GeForce® GTX 660 或更高配置 |
-| **内存** | 4 GB | 6 GB |
-| **存储** | 30 GB 可用的磁盘空间 | |
-| **网络** | 宽带连接 | |
-| **分辨率** | 1024 x 768 以上 | |
+## 验证安装
+
+以下示例基于 TensorFlow 1.x 。确保运行环境中使用的的是 ** TensorFlow 1.x**。
+
+* 通过克隆源代码下载示例。
+
+ ```bash
+ git clone -b v1.3 https://github.com/Microsoft/nni.git
+ ```
+
+* 运行 MNIST 示例。
+
+ Linux 或 macOS
+
+ ```bash
+ nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
+ ```
+
+ Windows
+
+ ```bash
+ nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
+ ```
+
+* 在命令行中等待输出 `INFO: Successfully started experiment!`。 此消息表明 Experiment 已成功启动。 通过命令行输出的 `Web UI url` 来访问 Experiment 的界面。
+
+```text
+INFO: Starting restful server...
+INFO: Successfully started Restful server!
+INFO: Setting local config...
+INFO: Successfully set local config!
+INFO: Starting experiment...
+INFO: Successfully started experiment!
+-----------------------------------------------------------------------
+The experiment id is egchD4qy
+The Web UI urls are: http://223.255.255.1:8080 http://127.0.0.1:8080
+-----------------------------------------------------------------------
+
+You can use these commands to get more information about the experiment
+-----------------------------------------------------------------------
+ commands description
+
+1. nnictl experiment show show the information of experiments
+2. nnictl trial ls list all of trial jobs
+3. nnictl top monitor the status of running experiments
+4. nnictl log stderr show stderr log content
+5. nnictl log stdout show stdout log content
+6. nnictl stop stop an experiment
+7. nnictl trial kill kill a trial job by id
+8. nnictl --help get help information about nnictl
+-----------------------------------------------------------------------
+```
+
+* 在浏览器中打开 `Web UI url`,可看到下图的 Experiment 详细信息,以及所有的 Trial 任务。 查看[这里](../Tutorial/WebUI.md)的更多页面。
+
+
+
+
+
+## 系统需求
+
+由于程序变更,NNI 的最低配置会有所更改。
+
+### Linux
+
+| | 推荐配置 | 最低配置 |
+| -------- | ----------------------------------------- | ------------------------------------- |
+| **操作系统** | Ubuntu 16.04 或以上版本 | |
+| **CPU** | Intel® Core™ i5 或 AMD Phenom™ II X3 或更高配置 | Intel® Core™ i3 或 AMD Phenom™ X3 8650 |
+| **GPU** | NVIDIA® GeForce® GTX 660 或更高配置 | NVIDIA® GeForce® GTX 460 |
+| **内存** | 6 GB | 4 GB |
+| **存储** | 30 GB 可用的磁盘空间 | |
+| **网络** | 宽带连接 | |
+| **分辨率** | 1024 x 768 以上 | |
+
+### macOS
+
+| | 推荐配置 | 最低配置 |
+| -------- | ------------------------ | -------------------------------------------------- |
+| **操作系统** | macOS 10.14.1 或更高版本 | |
+| **CPU** | Intel® Core™ i7-4770 或更高 | Intel® Core™ i5-760 或更高 |
+| **GPU** | AMD Radeon™ R9 M395X 或更高 | NVIDIA® GeForce® GT 750M 或 AMD Radeon™ R9 M290 或更高 |
+| **内存** | 8 GB | 4 GB |
+| **存储** | 70GB 可用空间 SSD 硬盘 | 70GB 可用空间及 7200 RPM 硬盘 |
+| **网络** | 宽带连接 | |
+| **分辨率** | 1024 x 768 以上 | |
+
+### Windows
+
+| | 推荐配置 | 最低配置 |
+| -------- | ----------------------------------------- | ------------------------------------- |
+| **操作系统** | Windows 10 1809 或更高版本 | |
+| **CPU** | Intel® Core™ i5 或 AMD Phenom™ II X3 或更高配置 | Intel® Core™ i3 或 AMD Phenom™ X3 8650 |
+| **GPU** | NVIDIA® GeForce® GTX 660 或更高配置 | NVIDIA® GeForce® GTX 460 |
+| **内存** | 6 GB | 4 GB |
+| **存储** | 30 GB 可用的磁盘空间 | |
+| **网络** | 宽带连接 | |
+| **分辨率** | 1024 x 768 以上 | |
## 更多
diff --git a/docs/zh_CN/Tutorial/Nnictl.md b/docs/zh_CN/Tutorial/Nnictl.md
index acee5d4534..38b66d314b 100644
--- a/docs/zh_CN/Tutorial/Nnictl.md
+++ b/docs/zh_CN/Tutorial/Nnictl.md
@@ -49,6 +49,7 @@ nnictl 支持的命令:
| --config, -c | True | | Experiment 的 YAML 配置文件 |
| --port, -p | False | | RESTful 服务的端口 |
| --debug, -d | False | | 设置为调试模式 |
+ | --watch, -w | False | | 启动为监视模式 |
* 示例
@@ -97,6 +98,7 @@ nnictl 支持的命令:
| id | True | | 要恢复的 Experiment 标识 |
| --port, -p | False | | 要恢复的 Experiment 使用的 RESTful 服务端口 |
| --debug, -d | False | | 设置为调试模式 |
+ | --watch, -w | False | | 启动为监视模式 |
* 示例
diff --git a/docs/zh_CN/Tutorial/QuickStart.md b/docs/zh_CN/Tutorial/QuickStart.md
index 3ed05f3e68..b886debf18 100644
--- a/docs/zh_CN/Tutorial/QuickStart.md
+++ b/docs/zh_CN/Tutorial/QuickStart.md
@@ -2,15 +2,15 @@
## 安装
-当前支持 Linux,MacOS 和 Windows,在 Ubuntu 16.04 或更高版本,MacOS 10.14.1 以及 Windows 10.1809 上进行了测试。 在 `python >= 3.5` 的环境中,只需要运行 `pip install` 即可完成安装。
+当前支持 Linux,macOS 和 Windows,在 Ubuntu 16.04 或更高版本,macOS 10.14.1 以及 Windows 10.1809 上进行了测试。 在 `python >= 3.5` 的环境中,只需要运行 `pip install` 即可完成安装。
-#### Linux 和 MacOS
+**Linux 和 macOS**
```bash
python3 -m pip install --upgrade nni
```
-#### Windows
+**Windows**
```bash
python -m pip install --upgrade nni
@@ -18,7 +18,7 @@
注意:
-* 在 Linux 和 MacOS 上,如果要将 NNI 安装到当前用户的 home 目录中,可使用 `--user`,则不需要特殊权限。
+* 在 Linux 和 macOS 上,如果要将 NNI 安装到当前用户的 home 目录中,可使用 `--user`,则不需要特殊权限。
* 如果遇到如`Segmentation fault` 这样的任何错误请参考[常见问题](FAQ.md)。
* 参考[安装 NNI](Installation.md),来了解`系统需求`。
@@ -54,21 +54,22 @@ if __name__ == '__main__':
NNI 用来帮助超参调优。它的流程如下:
- 输入: 搜索空间, Trial 代码, 配置文件
- 输出: 一组最佳的超参配置
-
- 1: For t = 0, 1, 2, ..., maxTrialNum,
- 2: hyperparameter = 从搜索空间选择一组参数
- 3: final result = run_trial_and_evaluate(hyperparameter)
- 4: 返回最终结果给 NNI
- 5: If 时间达到上限,
- 6: 停止实验
- 7: return 最好的实验结果
-
+```text
+输入: 搜索空间, Trial 代码, 配置文件
+输出: 一组最佳的超参配置
+
+1: For t = 0, 1, 2, ..., maxTrialNum,
+2: hyperparameter = 从搜索空间选择一组参数
+3: final result = run_trial_and_evaluate(hyperparameter)
+4: 返回最终结果给 NNI
+5: If 时间达到上限,
+6: 停止实验
+7: return 最好的实验结果
+```
如果需要使用 NNI 来自动训练模型,找到最佳超参,需要如下三步:
-**使用 NNI 时的三个步骤**
+**启动 Experiment 的三个步骤**
**第一步**:定义 JSON 格式的`搜索空间`文件,包括所有需要搜索的超参的`名称`和`分布`(离散和连续值均可)。
@@ -140,7 +141,7 @@ trial:
上面的代码都已准备好,并保存在 [examples/trials/mnist-tfv1/](https://github.com/Microsoft/nni/tree/master/examples/trials/mnist-tfv1)。
-#### Linux 和 macOS
+**Linux 和 macOS**
从命令行使用 **config.yml** 文件启动 MNIST Experiment 。
@@ -148,17 +149,17 @@ trial:
nnictl create --config nni/examples/trials/mnist-tfv1/config.yml
```
-#### Windows
+**Windows**
从命令行使用 **config_windows.yml** 文件启动 MNIST Experiment 。
-**注意**:如果使用 Windows,则需要在 config.yml 文件中,将 `python3` 改为 `python`,或者使用 config_windows.yml 来开始 Experiment。
+注意:如果使用 Windows,则需要在 config.yml 文件中,将 `python3` 改为 `python`,或者使用 config_windows.yml 来开始 Experiment。
```bash
nnictl create --config nni\examples\trials\mnist-tfv1\config_windows.yml
```
-注意:**nnictl** 是一个命令行工具,用来控制 NNI Experiment,如启动、停止、继续 Experiment,启动、停止 NNIBoard 等等。 查看[这里](Nnictl.md),了解 `nnictl` 更多用法。
+注意:`nnictl` 是一个命令行工具,用来控制 NNI Experiment,如启动、停止、继续 Experiment,启动、停止 NNIBoard 等等。 查看[这里](Nnictl.md),了解 `nnictl` 更多用法。
在命令行中等待输出 `INFO: Successfully started experiment!`。 此消息表明 Experiment 已成功启动。 期望的输出如下:
@@ -201,7 +202,7 @@ Web 地址为:[IP 地址]:8080
在浏览器中打开 `Web 界面地址`(即:`[IP 地址]:8080`),就可以看到 Experiment 的详细信息,以及所有的 Trial 任务。 如果无法打开终端中的 Web 界面链接,可以参考 [FAQ](FAQ.md)。
-#### 查看概要页面
+### 查看概要页面
点击标签 "Overview"。
@@ -213,7 +214,7 @@ Experiment 相关信息会显示在界面上,配置和搜索空间等。 可

-#### 查看 Trial 详情页面
+### 查看 Trial 详情页面
点击 "Default Metric" 来查看所有 Trial 的点图。 悬停鼠标来查看默认指标和搜索空间信息。
diff --git a/docs/zh_CN/conf.py b/docs/zh_CN/conf.py
index f1336f1c78..d5bec553af 100644
--- a/docs/zh_CN/conf.py
+++ b/docs/zh_CN/conf.py
@@ -47,6 +47,9 @@
'sphinx.ext.napoleon',
]
+# 添加示例模块
+autodoc_mock_imports = ['apex']
+
# Add any paths that contain templates here, relative to this directory.
templates_path = ['_templates']
@@ -72,7 +75,7 @@
# List of patterns, relative to source directory, that match files and
# directories to ignore when looking for source files.
# This pattern also affects html_static_path and html_extra_path.
-exclude_patterns = ['_build', 'Thumbs.db', '.DS_Store']
+exclude_patterns = ['_build', 'Thumbs.db', '.DS_Store', 'Release_v1.0.md']
# The name of the Pygments (syntax highlighting) style to use.
pygments_style = None
diff --git a/docs/zh_CN/examples.rst b/docs/zh_CN/examples.rst
index f76ce9eb3d..95e0f94fee 100644
--- a/docs/zh_CN/examples.rst
+++ b/docs/zh_CN/examples.rst
@@ -11,3 +11,5 @@
EvolutionSQuAD<./TrialExample/SquadEvolutionExamples>
GBDT<./TrialExample/GbdtExample>
RocksDB <./TrialExample/RocksdbExamples>
+ KD 示例 <./TrialExample/KDExample>
+ EfficientNet <./TrialExample/EfficientNet>
diff --git a/docs/zh_CN/model_compression.rst b/docs/zh_CN/model_compression.rst
index 34d05b4844..2e273a79eb 100644
--- a/docs/zh_CN/model_compression.rst
+++ b/docs/zh_CN/model_compression.rst
@@ -18,7 +18,7 @@ NNI 中也内置了一些流程的模型压缩算法。
概述
Level Pruner
AGP Pruner
- L1Filter Pruner
+ L1Filter Pruner
Slim Pruner
Lottery Ticket Pruner
FPGM Pruner
diff --git a/docs/zh_CN/nas.rst b/docs/zh_CN/nas.rst
index a7329dd60d..611c5aefe2 100644
--- a/docs/zh_CN/nas.rst
+++ b/docs/zh_CN/nas.rst
@@ -22,4 +22,6 @@ NAS 算法
NAS 接口
ENAS
DARTS
- P-DARTS
+ P-DARTS
+ SPOS
+ CDARTS
diff --git a/docs/zh_CN/training_services.rst b/docs/zh_CN/training_services.rst
index 4e2969e597..8e75af2ae7 100644
--- a/docs/zh_CN/training_services.rst
+++ b/docs/zh_CN/training_services.rst
@@ -6,5 +6,6 @@ NNI 支持的训练平台介绍
本机<./TrainingService/LocalMode>
远程<./TrainingService/RemoteMachineMode>
OpenPAI<./TrainingService/PaiMode>
+ OpenPAI Yarn 模式<./TrainingService/PaiYarnMode>
Kubeflow<./TrainingService/KubeflowMode>
FrameworkController<./TrainingService/FrameworkControllerMode>
diff --git a/examples/feature_engineering/auto-feature-engineering/README_zh_CN.md b/examples/feature_engineering/auto-feature-engineering/README_zh_CN.md
index 55b50217cd..76cce132ff 100644
--- a/examples/feature_engineering/auto-feature-engineering/README_zh_CN.md
+++ b/examples/feature_engineering/auto-feature-engineering/README_zh_CN.md
@@ -1,8 +1,7 @@
-**NNI 中的自动特征工程**
-===
+ **NNI 中的自动特征工程** ===
-此[示例](https://github.com/SpongebBob/tabular_automl_NNI)在 NNI 中实现了自动特征工程。
+ 此[示例](https://github.com/SpongebBob/tabular_automl_NNI)在 NNI 中实现了自动特征工程。
-代码来自于贡献者。 谢谢可爱的贡献者!
+ 代码来自于贡献者。 谢谢可爱的贡献者!
-欢迎越来越多的人加入我们!
\ No newline at end of file
+ 欢迎越来越多的人加入我们!
diff --git a/examples/nas/cdarts/aux_head.py b/examples/nas/cdarts/aux_head.py
new file mode 100644
index 0000000000..9a67d09fec
--- /dev/null
+++ b/examples/nas/cdarts/aux_head.py
@@ -0,0 +1,102 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import torch.nn as nn
+
+
+class DistillHeadCIFAR(nn.Module):
+
+ def __init__(self, C, size, num_classes, bn_affine=False):
+ """assuming input size 8x8 or 16x16"""
+ super(DistillHeadCIFAR, self).__init__()
+ self.features = nn.Sequential(
+ nn.ReLU(),
+ nn.AvgPool2d(size, stride=2, padding=0, count_include_pad=False), # image size = 2 x 2 / 6 x 6
+ nn.Conv2d(C, 128, 1, bias=False),
+ nn.BatchNorm2d(128, affine=bn_affine),
+ nn.ReLU(),
+ nn.Conv2d(128, 768, 2, bias=False),
+ nn.BatchNorm2d(768, affine=bn_affine),
+ nn.ReLU()
+ )
+ self.classifier = nn.Linear(768, num_classes)
+ self.gap = nn.AdaptiveAvgPool2d(1)
+
+ def forward(self, x):
+ x = self.features(x)
+ x = self.gap(x)
+ x = self.classifier(x.view(x.size(0), -1))
+ return x
+
+
+class DistillHeadImagenet(nn.Module):
+
+ def __init__(self, C, size, num_classes, bn_affine=False):
+ """assuming input size 7x7 or 14x14"""
+ super(DistillHeadImagenet, self).__init__()
+ self.features = nn.Sequential(
+ nn.ReLU(),
+ nn.AvgPool2d(size, stride=2, padding=0, count_include_pad=False), # image size = 2 x 2 / 6 x 6
+ nn.Conv2d(C, 128, 1, bias=False),
+ nn.BatchNorm2d(128, affine=bn_affine),
+ nn.ReLU(),
+ nn.Conv2d(128, 768, 2, bias=False),
+ nn.BatchNorm2d(768, affine=bn_affine),
+ nn.ReLU()
+ )
+ self.classifier = nn.Linear(768, num_classes)
+ self.gap = nn.AdaptiveAvgPool2d(1)
+
+ def forward(self, x):
+ x = self.features(x)
+ x = self.gap(x)
+ x = self.classifier(x.view(x.size(0), -1))
+ return x
+
+
+class AuxiliaryHeadCIFAR(nn.Module):
+
+ def __init__(self, C, size=5, num_classes=10):
+ """assuming input size 8x8"""
+ super(AuxiliaryHeadCIFAR, self).__init__()
+ self.features = nn.Sequential(
+ nn.ReLU(inplace=True),
+ nn.AvgPool2d(5, stride=3, padding=0, count_include_pad=False), # image size = 2 x 2
+ nn.Conv2d(C, 128, 1, bias=False),
+ nn.BatchNorm2d(128),
+ nn.ReLU(inplace=True),
+ nn.Conv2d(128, 768, 2, bias=False),
+ nn.BatchNorm2d(768),
+ nn.ReLU(inplace=True)
+ )
+ self.classifier = nn.Linear(768, num_classes)
+
+ def forward(self, x):
+ x = self.features(x)
+ x = self.classifier(x.view(x.size(0), -1))
+ return x
+
+
+class AuxiliaryHeadImageNet(nn.Module):
+
+ def __init__(self, C, size=5, num_classes=1000):
+ """assuming input size 7x7"""
+ super(AuxiliaryHeadImageNet, self).__init__()
+ self.features = nn.Sequential(
+ nn.ReLU(inplace=True),
+ nn.AvgPool2d(size, stride=2, padding=0, count_include_pad=False),
+ nn.Conv2d(C, 128, 1, bias=False),
+ nn.BatchNorm2d(128),
+ nn.ReLU(inplace=True),
+ nn.Conv2d(128, 768, 2, bias=False),
+ # NOTE: This batchnorm was omitted in my earlier implementation due to a typo.
+ # Commenting it out for consistency with the experiments in the paper.
+ # nn.BatchNorm2d(768),
+ nn.ReLU(inplace=True)
+ )
+ self.classifier = nn.Linear(768, num_classes)
+
+ def forward(self, x):
+ x = self.features(x)
+ x = self.classifier(x.view(x.size(0), -1))
+ return x
diff --git a/examples/nas/cdarts/config.py b/examples/nas/cdarts/config.py
new file mode 100644
index 0000000000..f0200f39cd
--- /dev/null
+++ b/examples/nas/cdarts/config.py
@@ -0,0 +1,137 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import argparse
+from functools import partial
+
+
+def get_parser(name):
+ """ make default formatted parser """
+ parser = argparse.ArgumentParser(name, formatter_class=argparse.ArgumentDefaultsHelpFormatter)
+ # print default value always
+ parser.add_argument = partial(parser.add_argument, help=' ')
+ return parser
+
+
+class BaseConfig(argparse.Namespace):
+ def print_params(self, prtf=print):
+ prtf("")
+ prtf("Parameters:")
+ for attr, value in sorted(vars(self).items()):
+ prtf("{}={}".format(attr.upper(), value))
+ prtf("")
+
+ def as_markdown(self):
+ """ Return configs as markdown format """
+ text = "|name|value| \n|-|-| \n"
+ for attr, value in sorted(vars(self).items()):
+ text += "|{}|{}| \n".format(attr, value)
+
+ return text
+
+
+class SearchConfig(BaseConfig):
+ def build_parser(self):
+ parser = get_parser("Search config")
+ ########### basic settings ############
+ parser.add_argument('--dataset', default='cifar10', choices=['cifar10', 'cifar100', 'imagenet'])
+ parser.add_argument('--n_classes', type=int, default=10)
+ parser.add_argument('--stem_multiplier', type=int, default=3)
+ parser.add_argument('--init_channels', type=int, default=16)
+ parser.add_argument('--data_dir', type=str, default='data/cifar', help='cifar dataset')
+ parser.add_argument('--output_path', type=str, default='./outputs', help='')
+ parser.add_argument('--batch_size', type=int, default=128, help='batch size')
+ parser.add_argument('--log_frequency', type=int, default=10, help='print frequency')
+ parser.add_argument('--seed', type=int, default=0, help='random seed')
+ parser.add_argument('--workers', type=int, default=4, help='# of workers')
+ parser.add_argument('--steps_per_epoch', type=int, default=None, help='how many steps per epoch, use None for one pass of dataset')
+
+ ########### learning rate ############
+ parser.add_argument('--w_lr', type=float, default=0.05, help='lr for weights')
+ parser.add_argument('--w_momentum', type=float, default=0.9, help='momentum for weights')
+ parser.add_argument('--w_weight_decay', type=float, default=3e-4, help='weight decay for weights')
+ parser.add_argument('--grad_clip', type=float, default=5., help='gradient clipping for weights')
+ parser.add_argument('--alpha_lr', type=float, default=6e-4, help='lr for alpha')
+ parser.add_argument('--alpha_weight_decay', type=float, default=1e-3, help='weight decay for alpha')
+ parser.add_argument('--nasnet_lr', type=float, default=0.1, help='lr of nasnet')
+
+ ########### alternate training ############
+ parser.add_argument('--epochs', type=int, default=32, help='# of search epochs')
+ parser.add_argument('--warmup_epochs', type=int, default=2, help='# warmup epochs of super model')
+ parser.add_argument('--loss_alpha', type=float, default=1, help='loss alpha')
+ parser.add_argument('--loss_T', type=float, default=2, help='loss temperature')
+ parser.add_argument('--interactive_type', type=str, default='kl', choices=['kl', 'smoothl1'])
+ parser.add_argument('--sync_bn', action='store_true', default=False, help='whether to sync bn')
+ parser.add_argument('--use_apex', action='store_true', default=False, help='whether to use apex')
+ parser.add_argument('--regular_ratio', type=float, default=0.5, help='regular ratio')
+ parser.add_argument('--regular_coeff', type=float, default=5, help='regular coefficient')
+ parser.add_argument('--fix_head', action='store_true', default=False, help='whether to fix head')
+ parser.add_argument('--share_module', action='store_true', default=False, help='whether to share stem and aux head')
+
+ ########### data augument ############
+ parser.add_argument('--aux_weight', type=float, default=0.4, help='auxiliary loss weight')
+ parser.add_argument('--cutout_length', type=int, default=16, help='cutout length')
+ parser.add_argument('--drop_path_prob', type=float, default=0.2, help='drop path prob')
+ parser.add_argument('--use_aa', action='store_true', default=False, help='whether to use aa')
+ parser.add_argument('--mixup_alpha', default=1., type=float, help='mixup interpolation coefficient (default: 1)')
+
+ ########### distributed ############
+ parser.add_argument("--local_rank", default=0, type=int)
+ parser.add_argument("--world_size", default=1, type=int)
+ parser.add_argument('--dist_url', default='tcp://127.0.0.1:23456', type=str, help='url used to set up distributed training')
+ parser.add_argument('--distributed', action='store_true', help='run model distributed mode')
+
+ return parser
+
+ def __init__(self):
+ parser = self.build_parser()
+ args = parser.parse_args()
+ super().__init__(**vars(args))
+
+
+class RetrainConfig(BaseConfig):
+ def build_parser(self):
+ parser = get_parser("Retrain config")
+ parser.add_argument('--dataset', default="cifar10", choices=['cifar10', 'cifar100', 'imagenet'])
+ parser.add_argument('--data_dir', type=str, default='data/cifar', help='cifar dataset')
+ parser.add_argument('--output_path', type=str, default='./outputs', help='')
+ parser.add_argument("--arc_checkpoint", default="epoch_02.json")
+ parser.add_argument('--log_frequency', type=int, default=10, help='print frequency')
+
+ ########### model settings ############
+ parser.add_argument('--n_classes', type=int, default=10)
+ parser.add_argument('--input_channels', type=int, default=3)
+ parser.add_argument('--stem_multiplier', type=int, default=3)
+ parser.add_argument('--batch_size', type=int, default=128, help='batch size')
+ parser.add_argument('--eval_batch_size', type=int, default=500, help='batch size for validation')
+ parser.add_argument('--lr', type=float, default=0.025, help='lr for weights')
+ parser.add_argument('--momentum', type=float, default=0.9, help='momentum')
+ parser.add_argument('--grad_clip', type=float, default=5., help='gradient clipping for weights')
+ parser.add_argument('--weight_decay', type=float, default=5e-4, help='weight decay')
+ parser.add_argument('--epochs', type=int, default=600, help='# of training epochs')
+ parser.add_argument('--warmup_epochs', type=int, default=5, help='# warmup')
+ parser.add_argument('--init_channels', type=int, default=36)
+ parser.add_argument('--layers', type=int, default=20, help='# of layers')
+ parser.add_argument('--seed', type=int, default=0, help='random seed')
+ parser.add_argument('--workers', type=int, default=4, help='# of workers')
+ parser.add_argument('--aux_weight', type=float, default=0.4, help='auxiliary loss weight')
+ parser.add_argument('--cutout_length', type=int, default=16, help='cutout length')
+ parser.add_argument('--label_smooth', type=float, default=0.1, help='label smoothing')
+ parser.add_argument('--drop_path_prob', type=float, default=0.3, help='drop path prob')
+
+ ########### data augmentation ############
+ parser.add_argument('--use_aa', action='store_true', default=False, help='whether to use aa')
+ parser.add_argument('--mixup_alpha', default=1., type=float, help='mixup interpolation coefficient')
+
+ ########### distributed ############
+ parser.add_argument("--local_rank", default=0, type=int)
+ parser.add_argument("--world_size", default=1, type=int)
+ parser.add_argument('--dist_url', default='tcp://127.0.0.1:23456', type=str, help='url used to set up distributed training')
+ parser.add_argument('--distributed', action='store_true', help='run model distributed mode')
+
+ return parser
+
+ def __init__(self):
+ parser = self.build_parser()
+ args = parser.parse_args()
+ super().__init__(**vars(args))
diff --git a/examples/nas/cdarts/datasets/cifar.py b/examples/nas/cdarts/datasets/cifar.py
new file mode 100644
index 0000000000..493335f151
--- /dev/null
+++ b/examples/nas/cdarts/datasets/cifar.py
@@ -0,0 +1,111 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import numpy as np
+import torch
+import torchvision.datasets as dset
+import torchvision.transforms as transforms
+
+from datasets.data_utils import CIFAR10Policy, Cutout
+from datasets.data_utils import SubsetDistributedSampler
+
+
+def data_transforms_cifar(config, cutout=False):
+ CIFAR_MEAN = [0.49139968, 0.48215827, 0.44653124]
+ CIFAR_STD = [0.24703233, 0.24348505, 0.26158768]
+
+ if config.use_aa:
+ train_transform = transforms.Compose([
+ transforms.RandomCrop(32, padding=4, fill=128),
+ transforms.RandomHorizontalFlip(), CIFAR10Policy(),
+ transforms.ToTensor(),
+ transforms.Normalize(CIFAR_MEAN, CIFAR_STD),
+ ])
+ else:
+ train_transform = transforms.Compose([
+ transforms.RandomCrop(32, padding=4),
+ transforms.RandomHorizontalFlip(),
+ transforms.ToTensor(),
+ transforms.Normalize(CIFAR_MEAN, CIFAR_STD),
+ ])
+
+ if cutout:
+ train_transform.transforms.append(Cutout(config.cutout_length))
+
+ valid_transform = transforms.Compose([
+ transforms.ToTensor(),
+ transforms.Normalize(CIFAR_MEAN, CIFAR_STD),
+ ])
+ return train_transform, valid_transform
+
+
+def get_search_datasets(config):
+ dataset = config.dataset.lower()
+ if dataset == 'cifar10':
+ dset_cls = dset.CIFAR10
+ n_classes = 10
+ elif dataset == 'cifar100':
+ dset_cls = dset.CIFAR100
+ n_classes = 100
+ else:
+ raise Exception("Not support dataset!")
+
+ train_transform, valid_transform = data_transforms_cifar(config, cutout=False)
+ train_data = dset_cls(root=config.data_dir, train=True, download=True, transform=train_transform)
+ test_data = dset_cls(root=config.data_dir, train=False, download=True, transform=valid_transform)
+
+ num_train = len(train_data)
+ indices = list(range(num_train))
+ split_mid = int(np.floor(0.5 * num_train))
+
+ if config.distributed:
+ train_sampler = SubsetDistributedSampler(train_data, indices[:split_mid])
+ valid_sampler = SubsetDistributedSampler(train_data, indices[split_mid:num_train])
+ else:
+ train_sampler = torch.utils.data.sampler.SubsetRandomSampler(indices[:split_mid])
+ valid_sampler = torch.utils.data.sampler.SubsetRandomSampler(indices[split_mid:num_train])
+
+ train_loader = torch.utils.data.DataLoader(
+ train_data, batch_size=config.batch_size,
+ sampler=train_sampler,
+ pin_memory=False, num_workers=config.workers)
+
+ valid_loader = torch.utils.data.DataLoader(
+ train_data, batch_size=config.batch_size,
+ sampler=valid_sampler,
+ pin_memory=False, num_workers=config.workers)
+
+ return [train_loader, valid_loader], [train_sampler, valid_sampler]
+
+
+def get_augment_datasets(config):
+ dataset = config.dataset.lower()
+ if dataset == 'cifar10':
+ dset_cls = dset.CIFAR10
+ elif dataset == 'cifar100':
+ dset_cls = dset.CIFAR100
+ else:
+ raise Exception("Not support dataset!")
+
+ train_transform, valid_transform = data_transforms_cifar(config, cutout=True)
+ train_data = dset_cls(root=config.data_dir, train=True, download=True, transform=train_transform)
+ test_data = dset_cls(root=config.data_dir, train=False, download=True, transform=valid_transform)
+
+ if config.distributed:
+ train_sampler = torch.utils.data.distributed.DistributedSampler(train_data)
+ test_sampler = torch.utils.data.distributed.DistributedSampler(test_data)
+ else:
+ train_sampler = None
+ test_sampler = None
+
+ train_loader = torch.utils.data.DataLoader(
+ train_data, batch_size=config.batch_size,
+ sampler=train_sampler,
+ pin_memory=True, num_workers=config.workers)
+
+ test_loader = torch.utils.data.DataLoader(
+ test_data, batch_size=config.eval_batch_size,
+ sampler=test_sampler,
+ pin_memory=True, num_workers=config.workers)
+
+ return [train_loader, test_loader], [train_sampler, test_sampler]
diff --git a/examples/nas/cdarts/datasets/data_utils.py b/examples/nas/cdarts/datasets/data_utils.py
new file mode 100644
index 0000000000..096b5a1fa7
--- /dev/null
+++ b/examples/nas/cdarts/datasets/data_utils.py
@@ -0,0 +1,400 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import math
+import random
+
+import numpy as np
+import torch
+import torch.distributed as dist
+from PIL import Image, ImageEnhance, ImageOps
+from torch.utils.data import Sampler
+
+
+class SubsetDistributedSampler(Sampler):
+ """
+ Sampler that restricts data loading to a subset of the dataset.
+
+ It is especially useful in conjunction with
+ :class:`torch.nn.parallel.DistributedDataParallel`. In such case, each
+ process can pass a DistributedSampler instance as a DataLoader sampler,
+ and load a subset of the original dataset that is exclusive to it.
+
+ Dataset is assumed to be of constant size.
+ """
+
+ def __init__(self, dataset, indices, num_replicas=None, rank=None, shuffle=True):
+ """
+ Initialization.
+
+ Parameters
+ ----------
+ dataset : torch.utils.data.Dataset
+ Dataset used for sampling.
+ num_replicas : int
+ Number of processes participating in distributed training. Default: World size.
+ rank : int
+ Rank of the current process within num_replicas. Default: Current rank.
+ shuffle : bool
+ If true (default), sampler will shuffle the indices.
+ """
+ if num_replicas is None:
+ if not dist.is_available():
+ raise RuntimeError("Requires distributed package to be available")
+ num_replicas = dist.get_world_size()
+ if rank is None:
+ if not dist.is_available():
+ raise RuntimeError("Requires distributed package to be available")
+ rank = dist.get_rank()
+ self.dataset = dataset
+ self.num_replicas = num_replicas
+ self.rank = rank
+ self.epoch = 0
+ self.indices = indices
+ self.num_samples = int(math.ceil(len(self.indices) * 1.0 / self.num_replicas))
+ self.total_size = self.num_samples * self.num_replicas
+ self.shuffle = shuffle
+
+ def __iter__(self):
+ # deterministically shuffle based on epoch
+ g = torch.Generator()
+ g.manual_seed(self.epoch)
+ if self.shuffle:
+ # indices = torch.randperm(len(self.dataset), generator=g).tolist()
+ indices = list(self.indices[i] for i in torch.randperm(len(self.indices)))
+ else:
+ # indices = list(range(len(self.dataset)))
+ indices = self.indices
+
+ # add extra samples to make it evenly divisible
+ indices += indices[:(self.total_size - len(indices))]
+ assert len(indices) == self.total_size
+
+ # subsample
+ indices = indices[self.rank:self.total_size:self.num_replicas]
+ assert len(indices) == self.num_samples
+
+ return iter(indices)
+
+ def __len__(self):
+ return self.num_samples
+
+ def set_epoch(self, epoch):
+ self.epoch = epoch
+
+
+class data_prefetcher():
+ def __init__(self, loader):
+ self.loader = iter(loader)
+ self.stream = torch.cuda.Stream()
+ self.mean = torch.tensor([0.485 * 255, 0.456 * 255, 0.406 * 255]).cuda().view(1, 3, 1, 1)
+ self.std = torch.tensor([0.229 * 255, 0.224 * 255, 0.225 * 255]).cuda().view(1, 3, 1, 1)
+ self.preload()
+
+ def preload(self):
+ try:
+ self.next_input, self.next_target = next(self.loader)
+ except StopIteration:
+ self.next_input = None
+ self.next_target = None
+ return
+ with torch.cuda.stream(self.stream):
+ self.next_input = self.next_input.cuda(non_blocking=True)
+ self.next_target = self.next_target.cuda(non_blocking=True)
+ self.next_input = self.next_input.float()
+ self.next_input = self.next_input.sub_(self.mean).div_(self.std)
+
+ def next(self):
+ torch.cuda.current_stream().wait_stream(self.stream)
+ input = self.next_input
+ target = self.next_target
+ self.preload()
+ return input, target
+
+
+class Cutout(object):
+ def __init__(self, length):
+ self.length = length
+
+ def __call__(self, img):
+ h, w = img.size(1), img.size(2)
+ mask = np.ones((h, w), np.float32)
+ y = np.random.randint(h)
+ x = np.random.randint(w)
+
+ y1 = np.clip(y - self.length // 2, 0, h)
+ y2 = np.clip(y + self.length // 2, 0, h)
+ x1 = np.clip(x - self.length // 2, 0, w)
+ x2 = np.clip(x + self.length // 2, 0, w)
+
+ mask[y1: y2, x1: x2] = 0.
+ mask = torch.from_numpy(mask)
+ mask = mask.expand_as(img)
+ img *= mask
+
+ return img
+
+
+class ImageNetPolicy(object):
+ """ Randomly choose one of the best 24 Sub-policies on ImageNet.
+ Example:
+ >>> policy = ImageNetPolicy()
+ >>> transformed = policy(image)
+ Example as a PyTorch Transform:
+ >>> transform=transforms.Compose([
+ >>> transforms.Resize(256),
+ >>> ImageNetPolicy(),
+ >>> transforms.ToTensor()])
+ """
+
+ def __init__(self, fillcolor=(128, 128, 128)):
+ self.policies = [
+ SubPolicy(0.4, "posterize", 8, 0.6, "rotate", 9, fillcolor),
+ SubPolicy(0.6, "solarize", 5, 0.6, "autocontrast", 5, fillcolor),
+ SubPolicy(0.8, "equalize", 8, 0.6, "equalize", 3, fillcolor),
+ SubPolicy(0.6, "posterize", 7, 0.6, "posterize", 6, fillcolor),
+ SubPolicy(0.4, "equalize", 7, 0.2, "solarize", 4, fillcolor),
+
+ SubPolicy(0.4, "equalize", 4, 0.8, "rotate", 8, fillcolor),
+ SubPolicy(0.6, "solarize", 3, 0.6, "equalize", 7, fillcolor),
+ SubPolicy(0.8, "posterize", 5, 1.0, "equalize", 2, fillcolor),
+ SubPolicy(0.2, "rotate", 3, 0.6, "solarize", 8, fillcolor),
+ SubPolicy(0.6, "equalize", 8, 0.4, "posterize", 6, fillcolor),
+
+ SubPolicy(0.8, "rotate", 8, 0.4, "color", 0, fillcolor),
+ SubPolicy(0.4, "rotate", 9, 0.6, "equalize", 2, fillcolor),
+ SubPolicy(0.0, "equalize", 7, 0.8, "equalize", 8, fillcolor),
+ SubPolicy(0.6, "invert", 4, 1.0, "equalize", 8, fillcolor),
+ SubPolicy(0.6, "color", 4, 1.0, "contrast", 8, fillcolor),
+
+ SubPolicy(0.8, "rotate", 8, 1.0, "color", 2, fillcolor),
+ SubPolicy(0.8, "color", 8, 0.8, "solarize", 7, fillcolor),
+ SubPolicy(0.4, "sharpness", 7, 0.6, "invert", 8, fillcolor),
+ SubPolicy(0.6, "shearX", 5, 1.0, "equalize", 9, fillcolor),
+ SubPolicy(0.4, "color", 0, 0.6, "equalize", 3, fillcolor),
+
+ SubPolicy(0.4, "equalize", 7, 0.2, "solarize", 4, fillcolor),
+ SubPolicy(0.6, "solarize", 5, 0.6, "autocontrast", 5, fillcolor),
+ SubPolicy(0.6, "invert", 4, 1.0, "equalize", 8, fillcolor),
+ SubPolicy(0.6, "color", 4, 1.0, "contrast", 8, fillcolor),
+ SubPolicy(0.8, "equalize", 8, 0.6, "equalize", 3, fillcolor)
+ ]
+
+ def __call__(self, img):
+ policy_idx = random.randint(0, len(self.policies) - 1)
+ return self.policies[policy_idx](img)
+
+ def __repr__(self):
+ return "AutoAugment ImageNet Policy"
+
+
+class CIFAR10Policy(object):
+ """ Randomly choose one of the best 25 Sub-policies on CIFAR10.
+ Example:
+ >>> policy = CIFAR10Policy()
+ >>> transformed = policy(image)
+ Example as a PyTorch Transform:
+ >>> transform=transforms.Compose([
+ >>> transforms.Resize(256),
+ >>> CIFAR10Policy(),
+ >>> transforms.ToTensor()])
+ """
+
+ def __init__(self, fillcolor=(128, 128, 128)):
+ self.policies = [
+ SubPolicy(0.1, "invert", 7, 0.2, "contrast", 6, fillcolor),
+ SubPolicy(0.7, "rotate", 2, 0.3, "translateX", 9, fillcolor),
+ SubPolicy(0.8, "sharpness", 1, 0.9, "sharpness", 3, fillcolor),
+ SubPolicy(0.5, "shearY", 8, 0.7, "translateY", 9, fillcolor),
+ SubPolicy(0.5, "autocontrast", 8, 0.9, "equalize", 2, fillcolor),
+
+ SubPolicy(0.2, "shearY", 7, 0.3, "posterize", 7, fillcolor),
+ SubPolicy(0.4, "color", 3, 0.6, "brightness", 7, fillcolor),
+ SubPolicy(0.3, "sharpness", 9, 0.7, "brightness", 9, fillcolor),
+ SubPolicy(0.6, "equalize", 5, 0.5, "equalize", 1, fillcolor),
+ SubPolicy(0.6, "contrast", 7, 0.6, "sharpness", 5, fillcolor),
+
+ SubPolicy(0.7, "color", 7, 0.5, "translateX", 8, fillcolor),
+ SubPolicy(0.3, "equalize", 7, 0.4, "autocontrast", 8, fillcolor),
+ SubPolicy(0.4, "translateY", 3, 0.2, "sharpness", 6, fillcolor),
+ SubPolicy(0.9, "brightness", 6, 0.2, "color", 8, fillcolor),
+ SubPolicy(0.5, "solarize", 2, 0.0, "invert", 3, fillcolor),
+
+ SubPolicy(0.2, "equalize", 0, 0.6, "autocontrast", 0, fillcolor),
+ SubPolicy(0.2, "equalize", 8, 0.6, "equalize", 4, fillcolor),
+ SubPolicy(0.9, "color", 9, 0.6, "equalize", 6, fillcolor),
+ SubPolicy(0.8, "autocontrast", 4, 0.2, "solarize", 8, fillcolor),
+ SubPolicy(0.1, "brightness", 3, 0.7, "color", 0, fillcolor),
+
+ SubPolicy(0.4, "solarize", 5, 0.9, "autocontrast", 3, fillcolor),
+ SubPolicy(0.9, "translateY", 9, 0.7, "translateY", 9, fillcolor),
+ SubPolicy(0.9, "autocontrast", 2, 0.8, "solarize", 3, fillcolor),
+ SubPolicy(0.8, "equalize", 8, 0.1, "invert", 3, fillcolor),
+ SubPolicy(0.7, "translateY", 9, 0.9, "autocontrast", 1, fillcolor)
+ ]
+
+ def __call__(self, img):
+ policy_idx = random.randint(0, len(self.policies) - 1)
+ return self.policies[policy_idx](img)
+
+ def __repr__(self):
+ return "AutoAugment CIFAR10 Policy"
+
+
+class SVHNPolicy(object):
+ """ Randomly choose one of the best 25 Sub-policies on SVHN.
+ Example:
+ >>> policy = SVHNPolicy()
+ >>> transformed = policy(image)
+ Example as a PyTorch Transform:
+ >>> transform=transforms.Compose([
+ >>> transforms.Resize(256),
+ >>> SVHNPolicy(),
+ >>> transforms.ToTensor()])
+ """
+
+ def __init__(self, fillcolor=(128, 128, 128)):
+ self.policies = [
+ SubPolicy(0.9, "shearX", 4, 0.2, "invert", 3, fillcolor),
+ SubPolicy(0.9, "shearY", 8, 0.7, "invert", 5, fillcolor),
+ SubPolicy(0.6, "equalize", 5, 0.6, "solarize", 6, fillcolor),
+ SubPolicy(0.9, "invert", 3, 0.6, "equalize", 3, fillcolor),
+ SubPolicy(0.6, "equalize", 1, 0.9, "rotate", 3, fillcolor),
+
+ SubPolicy(0.9, "shearX", 4, 0.8, "autocontrast", 3, fillcolor),
+ SubPolicy(0.9, "shearY", 8, 0.4, "invert", 5, fillcolor),
+ SubPolicy(0.9, "shearY", 5, 0.2, "solarize", 6, fillcolor),
+ SubPolicy(0.9, "invert", 6, 0.8, "autocontrast", 1, fillcolor),
+ SubPolicy(0.6, "equalize", 3, 0.9, "rotate", 3, fillcolor),
+
+ SubPolicy(0.9, "shearX", 4, 0.3, "solarize", 3, fillcolor),
+ SubPolicy(0.8, "shearY", 8, 0.7, "invert", 4, fillcolor),
+ SubPolicy(0.9, "equalize", 5, 0.6, "translateY", 6, fillcolor),
+ SubPolicy(0.9, "invert", 4, 0.6, "equalize", 7, fillcolor),
+ SubPolicy(0.3, "contrast", 3, 0.8, "rotate", 4, fillcolor),
+
+ SubPolicy(0.8, "invert", 5, 0.0, "translateY", 2, fillcolor),
+ SubPolicy(0.7, "shearY", 6, 0.4, "solarize", 8, fillcolor),
+ SubPolicy(0.6, "invert", 4, 0.8, "rotate", 4, fillcolor),
+ SubPolicy(0.3, "shearY", 7, 0.9, "translateX", 3, fillcolor),
+ SubPolicy(0.1, "shearX", 6, 0.6, "invert", 5, fillcolor),
+
+ SubPolicy(0.7, "solarize", 2, 0.6, "translateY", 7, fillcolor),
+ SubPolicy(0.8, "shearY", 4, 0.8, "invert", 8, fillcolor),
+ SubPolicy(0.7, "shearX", 9, 0.8, "translateY", 3, fillcolor),
+ SubPolicy(0.8, "shearY", 5, 0.7, "autocontrast", 3, fillcolor),
+ SubPolicy(0.7, "shearX", 2, 0.1, "invert", 5, fillcolor)
+ ]
+
+ def __call__(self, img):
+ policy_idx = random.randint(0, len(self.policies) - 1)
+ return self.policies[policy_idx](img)
+
+ def __repr__(self):
+ return "AutoAugment SVHN Policy"
+
+
+class SubPolicy(object):
+ def __init__(self, p1, operation1, magnitude_idx1, p2, operation2, magnitude_idx2, fillcolor=(128, 128, 128)):
+ ranges = {
+ "shearX": np.linspace(0, 0.3, 10),
+ "shearY": np.linspace(0, 0.3, 10),
+ "translateX": np.linspace(0, 150 / 331, 10),
+ "translateY": np.linspace(0, 150 / 331, 10),
+ "rotate": np.linspace(0, 30, 10),
+ "color": np.linspace(0.0, 0.9, 10),
+ "posterize": np.round(np.linspace(8, 4, 10), 0).astype(np.int),
+ "solarize": np.linspace(256, 0, 10),
+ "contrast": np.linspace(0.0, 0.9, 10),
+ "sharpness": np.linspace(0.0, 0.9, 10),
+ "brightness": np.linspace(0.0, 0.9, 10),
+ "autocontrast": [0] * 10,
+ "equalize": [0] * 10,
+ "invert": [0] * 10
+ }
+
+ # from https://stackoverflow.com/questions/5252170/specify-image-filling-color-when-rotating-in-python-with-pil-and-setting-expand
+ def rotate_with_fill(img, magnitude):
+ rot = img.convert("RGBA").rotate(magnitude)
+ return Image.composite(rot, Image.new("RGBA", rot.size, (128,) * 4), rot).convert(img.mode)
+
+ func = {
+ "shearX": lambda img, magnitude: img.transform(
+ img.size, Image.AFFINE, (1, magnitude * random.choice([-1, 1]), 0, 0, 1, 0),
+ Image.BICUBIC, fillcolor=fillcolor),
+ "shearY": lambda img, magnitude: img.transform(
+ img.size, Image.AFFINE, (1, 0, 0, magnitude * random.choice([-1, 1]), 1, 0),
+ Image.BICUBIC, fillcolor=fillcolor),
+ "translateX": lambda img, magnitude: img.transform(
+ img.size, Image.AFFINE, (1, 0, magnitude * img.size[0] * random.choice([-1, 1]), 0, 1, 0),
+ fillcolor=fillcolor),
+ "translateY": lambda img, magnitude: img.transform(
+ img.size, Image.AFFINE, (1, 0, 0, 0, 1, magnitude * img.size[1] * random.choice([-1, 1])),
+ fillcolor=fillcolor),
+ "rotate": lambda img, magnitude: rotate_with_fill(img, magnitude),
+ "color": lambda img, magnitude: ImageEnhance.Color(img).enhance(1 + magnitude * random.choice([-1, 1])),
+ "posterize": lambda img, magnitude: ImageOps.posterize(img, magnitude),
+ "solarize": lambda img, magnitude: ImageOps.solarize(img, magnitude),
+ "contrast": lambda img, magnitude: ImageEnhance.Contrast(img).enhance(
+ 1 + magnitude * random.choice([-1, 1])),
+ "sharpness": lambda img, magnitude: ImageEnhance.Sharpness(img).enhance(
+ 1 + magnitude * random.choice([-1, 1])),
+ "brightness": lambda img, magnitude: ImageEnhance.Brightness(img).enhance(
+ 1 + magnitude * random.choice([-1, 1])),
+ "autocontrast": lambda img, magnitude: ImageOps.autocontrast(img),
+ "equalize": lambda img, magnitude: ImageOps.equalize(img),
+ "invert": lambda img, magnitude: ImageOps.invert(img)
+ }
+
+ self.p1 = p1
+ self.operation1 = func[operation1]
+ self.magnitude1 = ranges[operation1][magnitude_idx1]
+ self.p2 = p2
+ self.operation2 = func[operation2]
+ self.magnitude2 = ranges[operation2][magnitude_idx2]
+
+ def __call__(self, img):
+ if random.random() < self.p1:
+ img = self.operation1(img, self.magnitude1)
+ if random.random() < self.p2:
+ img = self.operation2(img, self.magnitude2)
+ return img
+
+
+def fast_collate(batch):
+ imgs = [img[0] for img in batch]
+ targets = torch.tensor([target[1] for target in batch], dtype=torch.int64)
+ w = imgs[0].size[0]
+ h = imgs[0].size[1]
+ tensor = torch.zeros((len(imgs), 3, h, w), dtype=torch.uint8)
+ for i, img in enumerate(imgs):
+ nump_array = np.asarray(img, dtype=np.uint8)
+ if (nump_array.ndim < 3):
+ nump_array = np.expand_dims(nump_array, axis=-1)
+ nump_array = np.rollaxis(nump_array, 2)
+
+ tensor[i] += torch.from_numpy(nump_array)
+
+ return tensor, targets
+
+
+def mixup_data(x, y, alpha=1.0, use_cuda=True):
+ '''Returns mixed inputs, pairs of targets, and lambda'''
+ if alpha > 0:
+ lam = np.random.beta(alpha, alpha)
+ else:
+ lam = 1
+
+ batch_size = x.size()[0]
+ if use_cuda:
+ index = torch.randperm(batch_size).cuda()
+ else:
+ index = torch.randperm(batch_size)
+
+ mixed_x = lam * x + (1 - lam) * x[index, :]
+ y_a, y_b = y, y[index]
+ return mixed_x, y_a, y_b, lam
+
+
+def mixup_criterion(criterion, pred, y_a, y_b, lam):
+ return lam * criterion(pred, y_a) + (1 - lam) * criterion(pred, y_b)
diff --git a/examples/nas/cdarts/datasets/imagenet.py b/examples/nas/cdarts/datasets/imagenet.py
new file mode 100644
index 0000000000..3bba3d552e
--- /dev/null
+++ b/examples/nas/cdarts/datasets/imagenet.py
@@ -0,0 +1,100 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import os
+
+import numpy as np
+import torch
+import torchvision.datasets as dset
+import torchvision.transforms as transforms
+
+from datasets.data_utils import ImageNetPolicy
+from datasets.data_utils import SubsetDistributedSampler
+
+
+def _imagenet_dataset(config):
+ normalize = transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
+ train_dir = os.path.join(config.data_dir, "train")
+ test_dir = os.path.join(config.data_dir, "val")
+ if hasattr(config, "use_aa") and config.use_aa:
+ train_data = dset.ImageFolder(
+ train_dir,
+ transforms.Compose([
+ transforms.RandomResizedCrop(224),
+ transforms.RandomHorizontalFlip(),
+ ImageNetPolicy(),
+ transforms.ToTensor(),
+ normalize,
+ ]))
+ else:
+ train_data = dset.ImageFolder(
+ train_dir,
+ transforms.Compose([
+ transforms.RandomResizedCrop(224),
+ transforms.RandomHorizontalFlip(),
+ transforms.ColorJitter(
+ brightness=0.4,
+ contrast=0.4,
+ saturation=0.4,
+ hue=0.2),
+ transforms.ToTensor(),
+ normalize,
+ ]))
+
+ test_data = dset.ImageFolder(
+ test_dir,
+ transforms.Compose([
+ transforms.Resize(256),
+ transforms.CenterCrop(224),
+ transforms.ToTensor(),
+ normalize,
+ ]))
+
+ return train_data, test_data
+
+
+def get_search_datasets(config):
+ train_data, test_data = _imagenet_dataset(config)
+ num_train = len(train_data)
+ indices = list(range(num_train))
+ split_mid = int(np.floor(0.5 * num_train))
+
+ if config.distributed:
+ train_sampler = SubsetDistributedSampler(train_data, indices[:split_mid])
+ valid_sampler = SubsetDistributedSampler(train_data, indices[split_mid:num_train])
+ else:
+ train_sampler = torch.utils.data.sampler.SubsetRandomSampler(indices[:split_mid])
+ valid_sampler = torch.utils.data.sampler.SubsetRandomSampler(indices[split_mid:num_train])
+
+ train_loader = torch.utils.data.DataLoader(
+ train_data, batch_size=config.batch_size,
+ sampler=train_sampler,
+ pin_memory=True, num_workers=config.workers)
+
+ valid_loader = torch.utils.data.DataLoader(
+ train_data, batch_size=config.batch_size,
+ sampler=valid_sampler,
+ pin_memory=True, num_workers=config.workers)
+
+ return [train_loader, valid_loader], [train_sampler, valid_sampler]
+
+
+def get_augment_datasets(config):
+ train_data, test_data = _imagenet_dataset(config)
+ if config.distributed:
+ train_sampler = torch.utils.data.distributed.DistributedSampler(train_data)
+ test_sampler = torch.utils.data.distributed.DistributedSampler(test_data)
+ else:
+ train_sampler = test_sampler = None
+
+ train_loader = torch.utils.data.DataLoader(
+ train_data, batch_size=config.batch_size,
+ sampler=train_sampler,
+ pin_memory=True, num_workers=config.workers)
+
+ test_loader = torch.utils.data.DataLoader(
+ test_data, batch_size=config.batch_size,
+ sampler=test_sampler,
+ pin_memory=True, num_workers=config.workers)
+
+ return [train_loader, test_loader], [train_sampler, test_sampler]
diff --git a/examples/nas/cdarts/genotypes.py b/examples/nas/cdarts/genotypes.py
new file mode 100644
index 0000000000..0cc4d3fa63
--- /dev/null
+++ b/examples/nas/cdarts/genotypes.py
@@ -0,0 +1,166 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+"""
+- Genotype: normal/reduce gene + normal/reduce cell output connection (concat)
+- gene: discrete ops information (w/o output connection)
+- dag: real ops (can be mixed or discrete, but Genotype has only discrete information itself)
+"""
+from collections import namedtuple
+
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+
+import ops
+from ops import PRIMITIVES
+
+Genotype = namedtuple('Genotype', 'normal normal_concat reduce reduce_concat')
+
+
+def to_dag(C_in, gene, reduction, bn_affine=True):
+ """ generate discrete ops from gene """
+ dag = nn.ModuleList()
+ for edges in gene:
+ row = nn.ModuleList()
+ for op_name, s_idx in edges:
+ # reduction cell & from input nodes => stride = 2
+ stride = 2 if reduction and s_idx < 2 else 1
+ op = ops.OPS[op_name](C_in, stride, bn_affine)
+ if not isinstance(op, ops.Identity): # Identity does not use drop path
+ op = nn.Sequential(
+ op,
+ ops.DropPath_()
+ )
+ op.s_idx = s_idx
+ row.append(op)
+ dag.append(row)
+
+ return dag
+
+
+def from_str(s):
+ """ generate genotype from string
+ e.g. "Genotype(
+ normal=[[('sep_conv_3x3', 0), ('sep_conv_3x3', 1)],
+ [('sep_conv_3x3', 1), ('dil_conv_3x3', 2)],
+ [('sep_conv_3x3', 1), ('sep_conv_3x3', 2)],
+ [('sep_conv_3x3', 1), ('dil_conv_3x3', 4)]],
+ normal_concat=range(2, 6),
+ reduce=[[('max_pool_3x3', 0), ('max_pool_3x3', 1)],
+ [('max_pool_3x3', 0), ('skip_connect', 2)],
+ [('max_pool_3x3', 0), ('skip_connect', 2)],
+ [('max_pool_3x3', 0), ('skip_connect', 2)]],
+ reduce_concat=range(2, 6))"
+ """
+
+ genotype = eval(s)
+
+ return genotype
+
+
+def parse(alpha, beta, k):
+ """
+ parse continuous alpha to discrete gene.
+ alpha is ParameterList:
+ ParameterList [
+ Parameter(n_edges1, n_ops),
+ Parameter(n_edges2, n_ops),
+ ...
+ ]
+
+ beta is ParameterList:
+ ParameterList [
+ Parameter(n_edges1),
+ Parameter(n_edges2),
+ ...
+ ]
+
+ gene is list:
+ [
+ [('node1_ops_1', node_idx), ..., ('node1_ops_k', node_idx)],
+ [('node2_ops_1', node_idx), ..., ('node2_ops_k', node_idx)],
+ ...
+ ]
+ each node has two edges (k=2) in CNN.
+ """
+
+ gene = []
+ assert PRIMITIVES[-1] == 'none' # 'none' is implemented in mutator now
+
+ # 1) Convert the mixed op to discrete edge (single op) by choosing top-1 weight edge
+ # 2) Choose top-k edges per node by edge score (top-1 weight in edge)
+ # output the connect idx[(node_idx, connect_idx, op_idx).... () ()]
+ connect_idx = []
+ for edges, w in zip(alpha, beta):
+ # edges: Tensor(n_edges, n_ops)
+ edge_max, primitive_indices = torch.topk((w.view(-1, 1) * edges)[:, :-1], 1) # ignore 'none'
+ topk_edge_values, topk_edge_indices = torch.topk(edge_max.view(-1), k)
+ node_gene = []
+ node_idx = []
+ for edge_idx in topk_edge_indices:
+ prim_idx = primitive_indices[edge_idx]
+ prim = PRIMITIVES[prim_idx]
+ node_gene.append((prim, edge_idx.item()))
+ node_idx.append((edge_idx.item(), prim_idx.item()))
+
+ gene.append(node_gene)
+ connect_idx.append(node_idx)
+
+ return gene, connect_idx
+
+
+def parse_gumbel(alpha, beta, k):
+ """
+ parse continuous alpha to discrete gene.
+ alpha is ParameterList:
+ ParameterList [
+ Parameter(n_edges1, n_ops),
+ Parameter(n_edges2, n_ops),
+ ...
+ ]
+
+ beta is ParameterList:
+ ParameterList [
+ Parameter(n_edges1),
+ Parameter(n_edges2),
+ ...
+ ]
+
+ gene is list:
+ [
+ [('node1_ops_1', node_idx), ..., ('node1_ops_k', node_idx)],
+ [('node2_ops_1', node_idx), ..., ('node2_ops_k', node_idx)],
+ ...
+ ]
+ each node has two edges (k=2) in CNN.
+ """
+
+ gene = []
+ assert PRIMITIVES[-1] == 'none' # assume last PRIMITIVE is 'none'
+
+ # 1) Convert the mixed op to discrete edge (single op) by choosing top-1 weight edge
+ # 2) Choose top-k edges per node by edge score (top-1 weight in edge)
+ # output the connect idx[(node_idx, connect_idx, op_idx).... () ()]
+ connect_idx = []
+ for edges, w in zip(alpha, beta):
+ # edges: Tensor(n_edges, n_ops)
+ discrete_a = F.gumbel_softmax(edges[:, :-1].reshape(-1), tau=1, hard=True)
+ for i in range(k-1):
+ discrete_a = discrete_a + F.gumbel_softmax(edges[:, :-1].reshape(-1), tau=1, hard=True)
+ discrete_a = discrete_a.reshape(-1, len(PRIMITIVES)-1)
+ reserved_edge = (discrete_a > 0).nonzero()
+
+ node_gene = []
+ node_idx = []
+ for i in range(reserved_edge.shape[0]):
+ edge_idx = reserved_edge[i][0].item()
+ prim_idx = reserved_edge[i][1].item()
+ prim = PRIMITIVES[prim_idx]
+ node_gene.append((prim, edge_idx))
+ node_idx.append((edge_idx, prim_idx))
+
+ gene.append(node_gene)
+ connect_idx.append(node_idx)
+
+ return gene, connect_idx
diff --git a/examples/nas/cdarts/model.py b/examples/nas/cdarts/model.py
new file mode 100644
index 0000000000..0514004a5e
--- /dev/null
+++ b/examples/nas/cdarts/model.py
@@ -0,0 +1,162 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+
+import ops
+import numpy as np
+from nni.nas.pytorch import mutables
+from utils import parse_results
+from aux_head import DistillHeadCIFAR, DistillHeadImagenet, AuxiliaryHeadCIFAR, AuxiliaryHeadImageNet
+
+
+class Node(nn.Module):
+ def __init__(self, node_id, num_prev_nodes, channels, num_downsample_connect):
+ super().__init__()
+ self.ops = nn.ModuleList()
+ choice_keys = []
+ for i in range(num_prev_nodes):
+ stride = 2 if i < num_downsample_connect else 1
+ choice_keys.append("{}_p{}".format(node_id, i))
+ self.ops.append(mutables.LayerChoice([ops.OPS[k](channels, stride, False) for k in ops.PRIMITIVES],
+ key=choice_keys[-1]))
+ self.drop_path = ops.DropPath()
+ self.input_switch = mutables.InputChoice(choose_from=choice_keys, n_chosen=2, key="{}_switch".format(node_id))
+
+ def forward(self, prev_nodes):
+ assert len(self.ops) == len(prev_nodes)
+ out = [op(node) for op, node in zip(self.ops, prev_nodes)]
+ out = [self.drop_path(o) if o is not None else None for o in out]
+ return self.input_switch(out)
+
+
+class Cell(nn.Module):
+
+ def __init__(self, n_nodes, channels_pp, channels_p, channels, reduction_p, reduction):
+ super().__init__()
+ self.reduction = reduction
+ self.n_nodes = n_nodes
+
+ # If previous cell is reduction cell, current input size does not match with
+ # output size of cell[k-2]. So the output[k-2] should be reduced by preprocessing.
+ if reduction_p:
+ self.preproc0 = ops.FactorizedReduce(channels_pp, channels, affine=False)
+ else:
+ self.preproc0 = ops.StdConv(channels_pp, channels, 1, 1, 0, affine=False)
+ self.preproc1 = ops.StdConv(channels_p, channels, 1, 1, 0, affine=False)
+
+ # generate dag
+ self.mutable_ops = nn.ModuleList()
+ for depth in range(2, self.n_nodes + 2):
+ self.mutable_ops.append(Node("{}_n{}".format("reduce" if reduction else "normal", depth),
+ depth, channels, 2 if reduction else 0))
+
+ def forward(self, s0, s1):
+ # s0, s1 are the outputs of previous previous cell and previous cell, respectively.
+ tensors = [self.preproc0(s0), self.preproc1(s1)]
+ for node in self.mutable_ops:
+ cur_tensor = node(tensors)
+ tensors.append(cur_tensor)
+
+ output = torch.cat(tensors[2:], dim=1)
+ return output
+
+
+class Model(nn.Module):
+
+ def __init__(self, dataset, n_layers, in_channels=3, channels=16, n_nodes=4, retrain=False, shared_modules=None):
+ super().__init__()
+ assert dataset in ["cifar10", "imagenet"]
+ self.dataset = dataset
+ self.input_size = 32 if dataset == "cifar" else 224
+ self.in_channels = in_channels
+ self.channels = channels
+ self.n_nodes = n_nodes
+ self.aux_size = {2 * n_layers // 3: self.input_size // 4}
+ if dataset == "cifar10":
+ self.n_classes = 10
+ self.aux_head_class = AuxiliaryHeadCIFAR if retrain else DistillHeadCIFAR
+ if not retrain:
+ self.aux_size = {n_layers // 3: 6, 2 * n_layers // 3: 6}
+ elif dataset == "imagenet":
+ self.n_classes = 1000
+ self.aux_head_class = AuxiliaryHeadImageNet if retrain else DistillHeadImagenet
+ if not retrain:
+ self.aux_size = {n_layers // 3: 6, 2 * n_layers // 3: 5}
+ self.n_layers = n_layers
+ self.aux_head = nn.ModuleDict()
+ self.ensemble_param = nn.Parameter(torch.rand(len(self.aux_size) + 1) / (len(self.aux_size) + 1)) \
+ if not retrain else None
+
+ stem_multiplier = 3 if dataset == "cifar" else 1
+ c_cur = stem_multiplier * self.channels
+ self.shared_modules = {} # do not wrap with ModuleDict
+ if shared_modules is not None:
+ self.stem = shared_modules["stem"]
+ else:
+ self.stem = nn.Sequential(
+ nn.Conv2d(in_channels, c_cur, 3, 1, 1, bias=False),
+ nn.BatchNorm2d(c_cur)
+ )
+ self.shared_modules["stem"] = self.stem
+
+ # for the first cell, stem is used for both s0 and s1
+ # [!] channels_pp and channels_p is output channel size, but c_cur is input channel size.
+ channels_pp, channels_p, c_cur = c_cur, c_cur, channels
+
+ self.cells = nn.ModuleList()
+ reduction_p, reduction = False, False
+ aux_head_count = 0
+ for i in range(n_layers):
+ reduction_p, reduction = reduction, False
+ if i in [n_layers // 3, 2 * n_layers // 3]:
+ c_cur *= 2
+ reduction = True
+
+ cell = Cell(n_nodes, channels_pp, channels_p, c_cur, reduction_p, reduction)
+ self.cells.append(cell)
+ c_cur_out = c_cur * n_nodes
+ if i in self.aux_size:
+ if shared_modules is not None:
+ self.aux_head[str(i)] = shared_modules["aux" + str(aux_head_count)]
+ else:
+ self.aux_head[str(i)] = self.aux_head_class(c_cur_out, self.aux_size[i], self.n_classes)
+ self.shared_modules["aux" + str(aux_head_count)] = self.aux_head[str(i)]
+ aux_head_count += 1
+ channels_pp, channels_p = channels_p, c_cur_out
+
+ self.gap = nn.AdaptiveAvgPool2d(1)
+ self.linear = nn.Linear(channels_p, self.n_classes)
+
+ def forward(self, x):
+ s0 = s1 = self.stem(x)
+ outputs = []
+
+ for i, cell in enumerate(self.cells):
+ s0, s1 = s1, cell(s0, s1)
+ if str(i) in self.aux_head:
+ outputs.append(self.aux_head[str(i)](s1))
+
+ out = self.gap(s1)
+ out = out.view(out.size(0), -1) # flatten
+ logits = self.linear(out)
+ outputs.append(logits)
+
+ if self.ensemble_param is None:
+ assert len(outputs) == 2
+ return outputs[1], outputs[0]
+ else:
+ em_output = torch.cat([(e * o) for e, o in zip(F.softmax(self.ensemble_param, dim=0), outputs)], 0)
+ return logits, em_output
+
+ def drop_path_prob(self, p):
+ for module in self.modules():
+ if isinstance(module, ops.DropPath):
+ module.p = p
+
+ def plot_genotype(self, results, logger):
+ genotypes = parse_results(results, self.n_nodes)
+ logger.info(genotypes)
+ return genotypes
diff --git a/examples/nas/cdarts/ops.py b/examples/nas/cdarts/ops.py
new file mode 100644
index 0000000000..285dc2998b
--- /dev/null
+++ b/examples/nas/cdarts/ops.py
@@ -0,0 +1,161 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import torch
+import torch.nn as nn
+
+OPS = {
+ 'avg_pool_3x3': lambda C, stride, affine: PoolWithoutBN('avg', C, 3, stride, 1, affine=affine),
+ 'max_pool_3x3': lambda C, stride, affine: PoolWithoutBN('max', C, 3, stride, 1, affine=affine),
+ 'skip_connect': lambda C, stride, affine: nn.Identity() if stride == 1 else FactorizedReduce(C, C, affine=affine),
+ 'sep_conv_3x3': lambda C, stride, affine: SepConv(C, C, 3, stride, 1, affine=affine),
+ 'sep_conv_5x5': lambda C, stride, affine: SepConv(C, C, 5, stride, 2, affine=affine),
+ 'sep_conv_7x7': lambda C, stride, affine: SepConv(C, C, 7, stride, 3, affine=affine),
+ 'dil_conv_3x3': lambda C, stride, affine: DilConv(C, C, 3, stride, 2, 2, affine=affine), # 5x5
+ 'dil_conv_5x5': lambda C, stride, affine: DilConv(C, C, 5, stride, 4, 2, affine=affine), # 9x9
+ 'conv_7x1_1x7': lambda C, stride, affine: FacConv(C, C, 7, stride, 3, affine=affine)
+}
+
+PRIMITIVES = [
+ 'max_pool_3x3',
+ 'avg_pool_3x3',
+ 'skip_connect', # identity
+ 'sep_conv_3x3',
+ 'sep_conv_5x5',
+ 'dil_conv_3x3',
+ 'dil_conv_5x5',
+]
+
+
+class DropPath(nn.Module):
+ def __init__(self, p=0.):
+ """
+ Drop path with probability.
+
+ Parameters
+ ----------
+ p : float
+ Probability of an path to be zeroed.
+ """
+ super().__init__()
+ self.p = p
+
+ def forward(self, x):
+ if self.training and self.p > 0.:
+ keep_prob = 1. - self.p
+ # per data point mask
+ mask = torch.zeros((x.size(0), 1, 1, 1), device=x.device).bernoulli_(keep_prob)
+ return x / keep_prob * mask
+
+ return x
+
+
+class PoolWithoutBN(nn.Module):
+ """
+ AvgPool or MaxPool with BN. `pool_type` must be `max` or `avg`.
+ """
+
+ def __init__(self, pool_type, C, kernel_size, stride, padding, affine=True):
+ super().__init__()
+ if pool_type.lower() == 'max':
+ self.pool = nn.MaxPool2d(kernel_size, stride, padding)
+ elif pool_type.lower() == 'avg':
+ self.pool = nn.AvgPool2d(kernel_size, stride, padding, count_include_pad=False)
+ else:
+ raise NotImplementedError("Pool doesn't support pooling type other than max and avg.")
+
+ def forward(self, x):
+ out = self.pool(x)
+ return out
+
+
+class StdConv(nn.Module):
+ """
+ Standard conv: ReLU - Conv - BN
+ """
+
+ def __init__(self, C_in, C_out, kernel_size, stride, padding, affine=True):
+ super().__init__()
+ self.net = nn.Sequential(
+ nn.ReLU(),
+ nn.Conv2d(C_in, C_out, kernel_size, stride, padding, bias=False),
+ nn.BatchNorm2d(C_out, affine=affine)
+ )
+
+ def forward(self, x):
+ return self.net(x)
+
+
+class FacConv(nn.Module):
+ """
+ Factorized conv: ReLU - Conv(Kx1) - Conv(1xK) - BN
+ """
+
+ def __init__(self, C_in, C_out, kernel_length, stride, padding, affine=True):
+ super().__init__()
+ self.net = nn.Sequential(
+ nn.ReLU(),
+ nn.Conv2d(C_in, C_in, (kernel_length, 1), stride, padding, bias=False),
+ nn.Conv2d(C_in, C_out, (1, kernel_length), stride, padding, bias=False),
+ nn.BatchNorm2d(C_out, affine=affine)
+ )
+
+ def forward(self, x):
+ return self.net(x)
+
+
+class DilConv(nn.Module):
+ """
+ (Dilated) depthwise separable conv.
+ ReLU - (Dilated) depthwise separable - Pointwise - BN.
+ If dilation == 2, 3x3 conv => 5x5 receptive field, 5x5 conv => 9x9 receptive field.
+ """
+
+ def __init__(self, C_in, C_out, kernel_size, stride, padding, dilation, affine=True):
+ super().__init__()
+ self.net = nn.Sequential(
+ nn.ReLU(),
+ nn.Conv2d(C_in, C_in, kernel_size, stride, padding, dilation=dilation, groups=C_in,
+ bias=False),
+ nn.Conv2d(C_in, C_out, 1, stride=1, padding=0, bias=False),
+ nn.BatchNorm2d(C_out, affine=affine)
+ )
+
+ def forward(self, x):
+ return self.net(x)
+
+
+class SepConv(nn.Module):
+ """
+ Depthwise separable conv.
+ DilConv(dilation=1) * 2.
+ """
+
+ def __init__(self, C_in, C_out, kernel_size, stride, padding, affine=True):
+ super().__init__()
+ self.net = nn.Sequential(
+ DilConv(C_in, C_in, kernel_size, stride, padding, dilation=1, affine=affine),
+ DilConv(C_in, C_out, kernel_size, 1, padding, dilation=1, affine=affine)
+ )
+
+ def forward(self, x):
+ return self.net(x)
+
+
+class FactorizedReduce(nn.Module):
+ """
+ Reduce feature map size by factorized pointwise (stride=2).
+ """
+
+ def __init__(self, C_in, C_out, affine=True):
+ super().__init__()
+ self.relu = nn.ReLU()
+ self.conv1 = nn.Conv2d(C_in, C_out // 2, 1, stride=2, padding=0, bias=False)
+ self.conv2 = nn.Conv2d(C_in, C_out // 2, 1, stride=2, padding=0, bias=False)
+ self.bn = nn.BatchNorm2d(C_out, affine=affine)
+
+ def forward(self, x):
+ x = self.relu(x)
+ out = torch.cat([self.conv1(x), self.conv2(x[:, :, 1:, 1:])], dim=1)
+ out = self.bn(out)
+ return out
diff --git a/examples/nas/cdarts/retrain.py b/examples/nas/cdarts/retrain.py
new file mode 100644
index 0000000000..4cd320d58c
--- /dev/null
+++ b/examples/nas/cdarts/retrain.py
@@ -0,0 +1,156 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import json
+import logging
+import os
+import time
+from argparse import ArgumentParser
+
+import torch
+import torch.nn as nn
+
+import apex # pylint: disable=import-error
+import datasets
+import utils
+from apex.parallel import DistributedDataParallel # pylint: disable=import-error
+from config import RetrainConfig
+from datasets.cifar import get_augment_datasets
+from model import Model
+from nni.nas.pytorch.fixed import apply_fixed_architecture
+from nni.nas.pytorch.utils import AverageMeterGroup
+
+
+def train(logger, config, train_loader, model, optimizer, criterion, epoch, main_proc):
+ meters = AverageMeterGroup()
+ cur_lr = optimizer.param_groups[0]["lr"]
+ if main_proc:
+ logger.info("Epoch %d LR %.6f", epoch, cur_lr)
+
+ model.train()
+ for step, (x, y) in enumerate(train_loader):
+ x, y = x.cuda(non_blocking=True), y.cuda(non_blocking=True)
+ optimizer.zero_grad()
+ logits, aux_logits = model(x)
+ loss = criterion(logits, y)
+ if config.aux_weight > 0.:
+ loss += config.aux_weight * criterion(aux_logits, y)
+ loss.backward()
+ nn.utils.clip_grad_norm_(model.parameters(), config.grad_clip)
+ optimizer.step()
+
+ prec1, prec5 = utils.accuracy(logits, y, topk=(1, 5))
+ metrics = {"prec1": prec1, "prec5": prec5, "loss": loss}
+ metrics = utils.reduce_metrics(metrics, config.distributed)
+ meters.update(metrics)
+
+ if main_proc and (step % config.log_frequency == 0 or step + 1 == len(train_loader)):
+ logger.info("Epoch [%d/%d] Step [%d/%d] %s", epoch + 1, config.epochs, step + 1, len(train_loader), meters)
+
+ if main_proc:
+ logger.info("Train: [%d/%d] Final Prec@1 %.4f Prec@5 %.4f", epoch + 1, config.epochs, meters.prec1.avg, meters.prec5.avg)
+
+
+def validate(logger, config, valid_loader, model, criterion, epoch, main_proc):
+ meters = AverageMeterGroup()
+ model.eval()
+
+ with torch.no_grad():
+ for step, (x, y) in enumerate(valid_loader):
+ x, y = x.cuda(non_blocking=True), y.cuda(non_blocking=True)
+ logits, _ = model(x)
+ loss = criterion(logits, y)
+ prec1, prec5 = utils.accuracy(logits, y, topk=(1, 5))
+ metrics = {"prec1": prec1, "prec5": prec5, "loss": loss}
+ metrics = utils.reduce_metrics(metrics, config.distributed)
+ meters.update(metrics)
+
+ if main_proc and (step % config.log_frequency == 0 or step + 1 == len(valid_loader)):
+ logger.info("Epoch [%d/%d] Step [%d/%d] %s", epoch + 1, config.epochs, step + 1, len(valid_loader), meters)
+
+ if main_proc:
+ logger.info("Train: [%d/%d] Final Prec@1 %.4f Prec@5 %.4f", epoch + 1, config.epochs, meters.prec1.avg, meters.prec5.avg)
+ return meters.prec1.avg, meters.prec5.avg
+
+
+def main():
+ config = RetrainConfig()
+ main_proc = not config.distributed or config.local_rank == 0
+ if config.distributed:
+ torch.cuda.set_device(config.local_rank)
+ torch.distributed.init_process_group(backend='nccl', init_method=config.dist_url,
+ rank=config.local_rank, world_size=config.world_size)
+ if main_proc:
+ os.makedirs(config.output_path, exist_ok=True)
+ if config.distributed:
+ torch.distributed.barrier()
+ logger = utils.get_logger(os.path.join(config.output_path, 'search.log'))
+ if main_proc:
+ config.print_params(logger.info)
+ utils.reset_seed(config.seed)
+
+ loaders, samplers = get_augment_datasets(config)
+ train_loader, valid_loader = loaders
+ train_sampler, valid_sampler = samplers
+
+ model = Model(config.dataset, config.layers, in_channels=config.input_channels, channels=config.init_channels, retrain=True).cuda()
+ if config.label_smooth > 0:
+ criterion = utils.CrossEntropyLabelSmooth(config.n_classes, config.label_smooth)
+ else:
+ criterion = nn.CrossEntropyLoss()
+
+ fixed_arc_path = os.path.join(config.output_path, config.arc_checkpoint)
+ with open(fixed_arc_path, "r") as f:
+ fixed_arc = json.load(f)
+ fixed_arc = utils.encode_tensor(fixed_arc, torch.device("cuda"))
+ genotypes = utils.parse_results(fixed_arc, n_nodes=4)
+ genotypes_dict = {i: genotypes for i in range(3)}
+ apply_fixed_architecture(model, fixed_arc_path)
+ param_size = utils.param_size(model, criterion, [3, 32, 32] if 'cifar' in config.dataset else [3, 224, 224])
+
+ if main_proc:
+ logger.info("Param size: %.6f", param_size)
+ logger.info("Genotype: %s", genotypes)
+
+ # change training hyper parameters according to cell type
+ if 'cifar' in config.dataset:
+ if param_size < 3.0:
+ config.weight_decay = 3e-4
+ config.drop_path_prob = 0.2
+ elif 3.0 < param_size < 3.5:
+ config.weight_decay = 3e-4
+ config.drop_path_prob = 0.3
+ else:
+ config.weight_decay = 5e-4
+ config.drop_path_prob = 0.3
+
+ if config.distributed:
+ apex.parallel.convert_syncbn_model(model)
+ model = DistributedDataParallel(model, delay_allreduce=True)
+
+ optimizer = torch.optim.SGD(model.parameters(), config.lr, momentum=config.momentum, weight_decay=config.weight_decay)
+ lr_scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, config.epochs, eta_min=1E-6)
+
+ best_top1 = best_top5 = 0.
+ for epoch in range(config.epochs):
+ drop_prob = config.drop_path_prob * epoch / config.epochs
+ if config.distributed:
+ model.module.drop_path_prob(drop_prob)
+ else:
+ model.drop_path_prob(drop_prob)
+ # training
+ if config.distributed:
+ train_sampler.set_epoch(epoch)
+ train(logger, config, train_loader, model, optimizer, criterion, epoch, main_proc)
+
+ # validation
+ top1, top5 = validate(logger, config, valid_loader, model, criterion, epoch, main_proc)
+ best_top1 = max(best_top1, top1)
+ best_top5 = max(best_top5, top5)
+ lr_scheduler.step()
+
+ logger.info("Final best Prec@1 = %.4f Prec@5 = %.4f", best_top1, best_top5)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/nas/cdarts/run_retrain_cifar.sh b/examples/nas/cdarts/run_retrain_cifar.sh
new file mode 100755
index 0000000000..c78fd78343
--- /dev/null
+++ b/examples/nas/cdarts/run_retrain_cifar.sh
@@ -0,0 +1,13 @@
+NGPUS=4
+SGPU=0
+EGPU=$[NGPUS+SGPU-1]
+GPU_ID=`seq -s , $SGPU $EGPU`
+CUDA_VISIBLE_DEVICES=$GPU_ID python -m torch.distributed.launch --nproc_per_node=$NGPUS retrain.py \
+ --dataset cifar10 --n_classes 10 --init_channels 36 --stem_multiplier 3 \
+ --arc_checkpoint 'epoch_31.json' \
+ --batch_size 128 --workers 1 --log_frequency 10 \
+ --world_size $NGPUS --weight_decay 5e-4 \
+ --distributed --dist_url 'tcp://127.0.0.1:26443' \
+ --lr 0.1 --warmup_epochs 0 --epochs 600 \
+ --cutout_length 16 --aux_weight 0.4 --drop_path_prob 0.3 \
+ --label_smooth 0.0 --mixup_alpha 0
diff --git a/examples/nas/cdarts/run_search_cifar.sh b/examples/nas/cdarts/run_search_cifar.sh
new file mode 100755
index 0000000000..64c6b04da4
--- /dev/null
+++ b/examples/nas/cdarts/run_search_cifar.sh
@@ -0,0 +1,14 @@
+NGPUS=4
+SGPU=0
+EGPU=$[NGPUS+SGPU-1]
+GPU_ID=`seq -s , $SGPU $EGPU`
+CUDA_VISIBLE_DEVICES=$GPU_ID python -m torch.distributed.launch --nproc_per_node=$NGPUS search.py \
+ --dataset cifar10 --n_classes 10 --init_channels 16 --stem_multiplier 3 \
+ --batch_size 64 --workers 1 --log_frequency 10 \
+ --distributed --world_size $NGPUS --dist_url 'tcp://127.0.0.1:23343' \
+ --regular_ratio 0.2 --regular_coeff 5 \
+ --loss_alpha 1 --loss_T 2 \
+ --w_lr 0.2 --alpha_lr 3e-4 --nasnet_lr 0.2 \
+ --w_weight_decay 0. --alpha_weight_decay 0. \
+ --share_module --interactive_type kl \
+ --warmup_epochs 2 --epochs 32
diff --git a/examples/nas/cdarts/search.py b/examples/nas/cdarts/search.py
new file mode 100644
index 0000000000..c41f7ce1ff
--- /dev/null
+++ b/examples/nas/cdarts/search.py
@@ -0,0 +1,49 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import logging
+import os
+import random
+import time
+
+import numpy as np
+import torch
+import torch.nn as nn
+
+import utils
+from config import SearchConfig
+from datasets.cifar import get_search_datasets
+from model import Model
+from nni.nas.pytorch.cdarts import CdartsTrainer
+
+if __name__ == "__main__":
+ config = SearchConfig()
+ main_proc = not config.distributed or config.local_rank == 0
+ if config.distributed:
+ torch.cuda.set_device(config.local_rank)
+ torch.distributed.init_process_group(backend='nccl', init_method=config.dist_url,
+ rank=config.local_rank, world_size=config.world_size)
+ if main_proc:
+ os.makedirs(config.output_path, exist_ok=True)
+ if config.distributed:
+ torch.distributed.barrier()
+ logger = utils.get_logger(os.path.join(config.output_path, 'search.log'))
+ if main_proc:
+ config.print_params(logger.info)
+ utils.reset_seed(config.seed)
+
+ loaders, samplers = get_search_datasets(config)
+ model_small = Model(config.dataset, 8).cuda()
+ if config.share_module:
+ model_large = Model(config.dataset, 20, shared_modules=model_small.shared_modules).cuda()
+ else:
+ model_large = Model(config.dataset, 20).cuda()
+
+ criterion = nn.CrossEntropyLoss()
+ trainer = CdartsTrainer(model_small, model_large, criterion, loaders, samplers, logger,
+ config.regular_coeff, config.regular_ratio, config.warmup_epochs, config.fix_head,
+ config.epochs, config.steps_per_epoch, config.loss_alpha, config.loss_T, config.distributed,
+ config.log_frequency, config.grad_clip, config.interactive_type, config.output_path,
+ config.w_lr, config.w_momentum, config.w_weight_decay, config.alpha_lr, config.alpha_weight_decay,
+ config.nasnet_lr, config.local_rank, config.share_module)
+ trainer.train()
diff --git a/examples/nas/cdarts/utils.py b/examples/nas/cdarts/utils.py
new file mode 100644
index 0000000000..11febc0beb
--- /dev/null
+++ b/examples/nas/cdarts/utils.py
@@ -0,0 +1,136 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import json
+import logging
+import os
+import random
+from collections import namedtuple
+
+import numpy as np
+import torch
+import torch.distributed as dist
+import torch.nn as nn
+
+from genotypes import Genotype
+from ops import PRIMITIVES
+from nni.nas.pytorch.cdarts.utils import *
+
+
+def get_logger(file_path):
+ """ Make python logger """
+ logger = logging.getLogger('cdarts')
+ log_format = '%(asctime)s | %(message)s'
+ formatter = logging.Formatter(log_format, datefmt='%m/%d %I:%M:%S %p')
+ file_handler = logging.FileHandler(file_path)
+ file_handler.setFormatter(formatter)
+ # stream_handler = logging.StreamHandler()
+ # stream_handler.setFormatter(formatter)
+
+ logger.addHandler(file_handler)
+ # logger.addHandler(stream_handler)
+ logger.setLevel(logging.INFO)
+
+ return logger
+
+
+class CyclicIterator:
+ def __init__(self, loader, sampler, distributed):
+ self.loader = loader
+ self.sampler = sampler
+ self.epoch = 0
+ self.distributed = distributed
+ self._next_epoch()
+
+ def _next_epoch(self):
+ if self.distributed:
+ self.sampler.set_epoch(self.epoch)
+ self.iterator = iter(self.loader)
+ self.epoch += 1
+
+ def __len__(self):
+ return len(self.loader)
+
+ def __iter__(self):
+ return self
+
+ def __next__(self):
+ try:
+ return next(self.iterator)
+ except StopIteration:
+ self._next_epoch()
+ return next(self.iterator)
+
+
+class CrossEntropyLabelSmooth(nn.Module):
+
+ def __init__(self, num_classes, epsilon):
+ super(CrossEntropyLabelSmooth, self).__init__()
+ self.num_classes = num_classes
+ self.epsilon = epsilon
+ self.logsoftmax = nn.LogSoftmax(dim=1)
+
+ def forward(self, inputs, targets):
+ log_probs = self.logsoftmax(inputs)
+ targets = torch.zeros_like(log_probs).scatter_(1, targets.unsqueeze(1), 1)
+ targets = (1 - self.epsilon) * targets + self.epsilon / self.num_classes
+ loss = (-targets * log_probs).mean(0).sum()
+ return loss
+
+def parse_results(results, n_nodes):
+ concat = range(2, 2 + n_nodes)
+ normal_gene = []
+ reduction_gene = []
+ for i in range(n_nodes):
+ normal_node = []
+ reduction_node = []
+ for j in range(2 + i):
+ normal_key = 'normal_n{}_p{}'.format(i + 2, j)
+ reduction_key = 'reduce_n{}_p{}'.format(i + 2, j)
+ normal_op = results[normal_key].cpu().numpy()
+ reduction_op = results[reduction_key].cpu().numpy()
+ if sum(normal_op == 1):
+ normal_index = np.argmax(normal_op)
+ normal_node.append((PRIMITIVES[normal_index], j))
+ if sum(reduction_op == 1):
+ reduction_index = np.argmax(reduction_op)
+ reduction_node.append((PRIMITIVES[reduction_index], j))
+ normal_gene.append(normal_node)
+ reduction_gene.append(reduction_node)
+
+ genotypes = Genotype(normal=normal_gene, normal_concat=concat,
+ reduce=reduction_gene, reduce_concat=concat)
+ return genotypes
+
+
+def param_size(model, loss_fn, input_size):
+ """
+ Compute parameter size in MB
+ """
+ x = torch.rand([2] + input_size).cuda()
+ y, _ = model(x)
+ target = torch.randint(model.n_classes, size=[2]).cuda()
+ loss = loss_fn(y, target)
+ loss.backward()
+ n_params = sum(np.prod(v.size()) for k, v in model.named_parameters() if not k.startswith('aux_head') and v.grad is not None)
+ return n_params / 1e6
+
+
+def encode_tensor(data, device):
+ if isinstance(data, list):
+ if all(map(lambda o: isinstance(o, bool), data)):
+ return torch.tensor(data, dtype=torch.bool, device=device) # pylint: disable=not-callable
+ else:
+ return torch.tensor(data, dtype=torch.float, device=device) # pylint: disable=not-callable
+ if isinstance(data, dict):
+ return {k: encode_tensor(v, device) for k, v in data.items()}
+ return data
+
+
+def reset_seed(seed):
+ torch.manual_seed(seed)
+ torch.cuda.manual_seed_all(seed)
+ np.random.seed(seed)
+ random.seed(seed)
+ torch.backends.cudnn.deterministic = True
+ torch.backends.cudnn.benchmark = True
diff --git a/examples/trials/efficientnet/README.md b/examples/trials/efficientnet/README.md
index 2d8f436594..0ed4a844e0 100644
--- a/examples/trials/efficientnet/README.md
+++ b/examples/trials/efficientnet/README.md
@@ -1,19 +1 @@
-# EfficientNet
-
-[EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks](https://arxiv.org/abs/1905.11946)
-
-Provided here are: Search space and tuners for finding the best tuple (alpha, beta, gamma) for EfficientNet-B1 with grid search, as discussed in Section 3.3 in [paper](https://arxiv.org/abs/1905.11946).
-
-## Instructions
-
-1. Set your working directory here in this directory.
-2. Run `git clone https://github.com/ultmaster/EfficientNet-PyTorch` to clone this modified version of [EfficientNet-PyTorch](https://github.com/lukemelas/EfficientNet-PyTorch). The modifications were done to adhere to the original [Tensorflow version](https://github.com/tensorflow/tpu/tree/master/models/official/efficientnet) as close as possible (including EMA, label smoothing and etc.); also added are the part which gets parameters from tuner and reports intermediate/final results. Clone it into `EfficientNet-PyTorch`; the files like `main.py`, `train_imagenet.sh` will appear inside, as specified in the configuration files.
-3. Run `nnictl create --config config_net.yml` to find the best EfficientNet-B1. Adjust the training service (PAI/local/remote), batch size in the config files according to the environment.
-
-For training on ImageNet, read `EfficientNet-PyTorch/train_imagenet.sh`. Download ImageNet beforehand and extract it adhering to [PyTorch format](https://pytorch.org/docs/stable/torchvision/datasets.html#imagenet) and then replace `/mnt/data/imagenet` in with the location of the ImageNet storage. This file should also be a good example to follow for mounting ImageNet into the container on OpenPAI.
-
-## Results
-
-The follow image is a screenshot, demonstrating the relationship between acc@1 and alpha, beta, gamma.
-
-
\ No newline at end of file
+[Documentation](https://nni.readthedocs.io/en/latest/TrialExample/EfficientNet.html)
diff --git a/examples/trials/efficientnet/README_zh_CN.md b/examples/trials/efficientnet/README_zh_CN.md
index 2f4ac5e65f..083689141b 100644
--- a/examples/trials/efficientnet/README_zh_CN.md
+++ b/examples/trials/efficientnet/README_zh_CN.md
@@ -1,19 +1 @@
-# EfficientNet
-
-[EfficientNet: 重新思考卷积神经网络的模型尺度](https://arxiv.org/abs/1905.11946)
-
-这里提供了:使用遍历搜索为 EfficientNet-B1 找到最佳元组(alpha,beta,gamma)的搜索空间和 Tuner。参考[论文](https://arxiv.org/abs/1905.11946) 3.3。
-
-## 说明
-
-1. 设置此目录为当前目录。
-2. 运行 `git clone https://github.com/ultmaster/EfficientNet-PyTorch` 来 clone 修改过的 [EfficientNet-PyTorch](https://github.com/lukemelas/EfficientNet-PyTorch)。 修改尽可能接近原始的 [TensorFlow 版本](https://github.com/tensorflow/tpu/tree/master/models/official/efficientnet) (包括 EMA,标记平滑度等等。);另外添加了代码从 Tuner 获取参数并回调中间和最终结果。 将其 clone 至 `EfficientNet-PyTorch`;`main.py`,`train_imagenet.sh` 等文件会在配置文件中指定的路径。
-3. 运行 `nnictl create --config config_net.yml` 来找到最好的 EfficientNet-B1。 根据环境来调整训练平台(OpenPAI、本机、远程),batch size。
-
-在 ImageNet 上的训练,可阅读 `EfficientNet-PyTorch/train_imagenet.sh`。 下载 ImageNet,并参考 [PyTorch 格式](https://pytorch.org/docs/stable/torchvision/datasets.html#imagenet) 来解压,然后将 `/mnt/data/imagenet` 替换为 ImageNet 的路径。 此文件也是如何将 ImageNet 挂载到 OpenPAI 容器的示例。
-
-## 结果
-
-下图展示了 acc@1 和 alpha、beta、gamma 之间的关系。
-
-
\ No newline at end of file
+[文档](https://nni.readthedocs.io/en/latest/TrialExample/EfficientNet.html)
\ No newline at end of file
diff --git a/examples/trials/efficientnet/config_local.yml b/examples/trials/efficientnet/config_local.yml
new file mode 100644
index 0000000000..bbb0978be2
--- /dev/null
+++ b/examples/trials/efficientnet/config_local.yml
@@ -0,0 +1,18 @@
+authorName: unknown
+experimentName: example_efficient_net
+trialConcurrency: 4
+maxExecDuration: 99999d
+maxTrialNum: 100
+trainingServicePlatform: local
+searchSpacePath: search_net.json
+useAnnotation: false
+tuner:
+ codeDir: .
+ classFileName: tuner.py
+ className: FixedProductTuner
+ classArgs:
+ product: 2
+trial:
+ codeDir: EfficientNet-PyTorch
+ command: python main.py /data/imagenet -j 12 -a efficientnet --batch-size 48 --lr 0.048 --wd 1e-5 --epochs 5 --request-from-nni
+ gpuNum: 1
diff --git a/examples/trials/efficientnet/config_net.yml b/examples/trials/efficientnet/config_pai.yml
similarity index 100%
rename from examples/trials/efficientnet/config_net.yml
rename to examples/trials/efficientnet/config_pai.yml
diff --git a/examples/trials/efficientnet/search_net.json b/examples/trials/efficientnet/search_net.json
index bf45ba918d..360441711f 100644
--- a/examples/trials/efficientnet/search_net.json
+++ b/examples/trials/efficientnet/search_net.json
@@ -1,14 +1,14 @@
{
"alpha": {
"_type": "quniform",
- "_value": [1.0, 2.0, 0.1]
+ "_value": [1.0, 2.0, 0.05]
},
"beta": {
"_type": "quniform",
- "_value": [1.0, 1.5, 0.1]
+ "_value": [1.0, 1.5, 0.05]
},
"gamma": {
"_type": "quniform",
- "_value": [1.0, 1.5, 0.1]
+ "_value": [1.0, 1.5, 0.05]
}
}
diff --git a/examples/trials/efficientnet/tuner.py b/examples/trials/efficientnet/tuner.py
index d091d40ac0..7e5bc8b60c 100644
--- a/examples/trials/efficientnet/tuner.py
+++ b/examples/trials/efficientnet/tuner.py
@@ -14,11 +14,11 @@ def __init__(self, product):
super().__init__()
self.product = product
- def expand_parameters(self, para):
+ def _expand_parameters(self, para):
"""
Filter out all qualified parameters
"""
- para = super().expand_parameters(para)
+ para = super()._expand_parameters(para)
if all([key in para[0] for key in ["alpha", "beta", "gamma"]]): # if this is an interested set
ret_para = []
for p in para:
diff --git a/src/nni_manager/core/nniDataStore.ts b/src/nni_manager/core/nniDataStore.ts
index 47134430d7..2d0dab8bee 100644
--- a/src/nni_manager/core/nniDataStore.ts
+++ b/src/nni_manager/core/nniDataStore.ts
@@ -4,6 +4,7 @@
'use strict';
import * as assert from 'assert';
+import * as JSON5 from 'json5';
import { Deferred } from 'ts-deferred';
import * as component from '../common/component';
@@ -131,7 +132,7 @@ class NNIDataStore implements DataStore {
}
public async storeMetricData(trialJobId: string, data: string): Promise {
- const metrics: MetricData = JSON.parse(data);
+ const metrics: MetricData = JSON5.parse(data);
// REQUEST_PARAMETER is used to request new parameters for multiphase trial job,
// it is not metrics, so it is skipped here.
if (metrics.type === 'REQUEST_PARAMETER') {
@@ -140,7 +141,7 @@ class NNIDataStore implements DataStore {
}
assert(trialJobId === metrics.trial_job_id);
try {
- await this.db.storeMetricData(trialJobId, JSON.stringify({
+ await this.db.storeMetricData(trialJobId, JSON5.stringify({
trialJobId: metrics.trial_job_id,
parameterId: metrics.parameter_id,
type: metrics.type,
diff --git a/src/nni_manager/core/sqlDatabase.ts b/src/nni_manager/core/sqlDatabase.ts
index 125a1aff6d..0ad6fd6bbe 100644
--- a/src/nni_manager/core/sqlDatabase.ts
+++ b/src/nni_manager/core/sqlDatabase.ts
@@ -5,6 +5,7 @@
import * as assert from 'assert';
import * as fs from 'fs';
+import * as JSON5 from 'json5';
import * as path from 'path';
import * as sqlite3 from 'sqlite3';
import { Deferred } from 'ts-deferred';
@@ -202,10 +203,10 @@ class SqlDB implements Database {
public storeMetricData(trialJobId: string, data: string): Promise {
const sql: string = 'insert into MetricData values (?,?,?,?,?,?)';
- const json: MetricDataRecord = JSON.parse(data);
- const args: any[] = [Date.now(), json.trialJobId, json.parameterId, json.type, json.sequence, JSON.stringify(json.data)];
+ const json: MetricDataRecord = JSON5.parse(data);
+ const args: any[] = [Date.now(), json.trialJobId, json.parameterId, json.type, json.sequence, JSON5.stringify(json.data)];
- this.log.trace(`storeMetricData: SQL: ${sql}, args: ${JSON.stringify(args)}`);
+ this.log.trace(`storeMetricData: SQL: ${sql}, args: ${JSON5.stringify(args)}`);
const deferred: Deferred = new Deferred();
this.db.run(sql, args, (err: Error | null) => { this.resolve(deferred, err); });
diff --git a/src/nni_manager/package.json b/src/nni_manager/package.json
index f79a0a4529..93e77cdf48 100644
--- a/src/nni_manager/package.json
+++ b/src/nni_manager/package.json
@@ -16,6 +16,7 @@
"express": "^4.16.3",
"express-joi-validator": "^2.0.0",
"js-base64": "^2.4.9",
+ "json5": "^2.1.1",
"kubernetes-client": "^6.5.0",
"rx": "^4.1.0",
"sqlite3": "^4.0.2",
@@ -34,6 +35,7 @@
"@types/express": "^4.16.0",
"@types/glob": "^7.1.1",
"@types/js-base64": "^2.3.1",
+ "@types/json5": "^0.0.30",
"@types/mocha": "^5.2.5",
"@types/node": "10.12.18",
"@types/request": "^2.47.1",
diff --git a/src/nni_manager/training_service/pai/paiK8S/paiK8STrainingService.ts b/src/nni_manager/training_service/pai/paiK8S/paiK8STrainingService.ts
index fc64d4dbdc..b6b6b4c823 100644
--- a/src/nni_manager/training_service/pai/paiK8S/paiK8STrainingService.ts
+++ b/src/nni_manager/training_service/pai/paiK8S/paiK8STrainingService.ts
@@ -59,6 +59,10 @@ class PAIK8STrainingService extends PAITrainingService {
public async setClusterMetadata(key: string, value: string): Promise {
switch (key) {
+ case TrialConfigMetadataKey.NNI_MANAGER_IP:
+ this.nniManagerIpConfig = JSON.parse(value);
+ break;
+
case TrialConfigMetadataKey.PAI_CLUSTER_CONFIG:
this.paiJobRestServer = new PAIJobRestServer(component.get(PAIK8STrainingService));
this.paiClusterConfig = JSON.parse(value);
diff --git a/src/nni_manager/training_service/pai/paiTrainingService.ts b/src/nni_manager/training_service/pai/paiTrainingService.ts
index 01cd4ed9dc..4f44dad8c6 100644
--- a/src/nni_manager/training_service/pai/paiTrainingService.ts
+++ b/src/nni_manager/training_service/pai/paiTrainingService.ts
@@ -151,18 +151,20 @@ abstract class PAITrainingService implements TrainingService {
public cancelTrialJob(trialJobId: string, isEarlyStopped: boolean = false): Promise {
const trialJobDetail: PAITrialJobDetail | undefined = this.trialJobsMap.get(trialJobId);
- const deferred: Deferred = new Deferred();
if (trialJobDetail === undefined) {
- this.log.error(`cancelTrialJob: trial job id ${trialJobId} not found`);
-
- return Promise.reject();
+ return Promise.reject(new Error(`cancelTrialJob: trial job id ${trialJobId} not found`));
}
if (this.paiClusterConfig === undefined) {
- throw new Error('PAI Cluster config is not initialized');
+ return Promise.reject(new Error('PAI Cluster config is not initialized'));
}
if (this.paiToken === undefined) {
- throw new Error('PAI token is not initialized');
+ return Promise.reject(new Error('PAI token is not initialized'));
+ }
+
+ if (trialJobDetail.status === 'UNKNOWN') {
+ trialJobDetail.status = 'USER_CANCELED';
+ return Promise.resolve();
}
const stopJobRequest: request.Options = {
@@ -179,6 +181,7 @@ abstract class PAITrainingService implements TrainingService {
// Set trialjobDetail's early stopped field, to mark the job's cancellation source
trialJobDetail.isEarlyStopped = isEarlyStopped;
+ const deferred: Deferred = new Deferred();
request(stopJobRequest, (error: Error, response: request.Response, body: any) => {
if ((error !== undefined && error !== null) || response.statusCode >= 400) {
diff --git a/src/nni_manager/training_service/remote_machine/remoteMachineTrainingService.ts b/src/nni_manager/training_service/remote_machine/remoteMachineTrainingService.ts
index bc0f44fc58..e88ae8aed2 100644
--- a/src/nni_manager/training_service/remote_machine/remoteMachineTrainingService.ts
+++ b/src/nni_manager/training_service/remote_machine/remoteMachineTrainingService.ts
@@ -277,6 +277,12 @@ class RemoteMachineTrainingService implements TrainingService {
throw new Error(`Invalid job id ${trialJobId}, cannot find ssh client`);
}
+ if (trialJob.status === 'UNKNOWN') {
+ this.releaseTrialSSHClient(trialJob);
+ trialJob.status = 'USER_CANCELED';
+ return
+ }
+
const jobpidPath: string = this.getJobPidPath(trialJob.id);
try {
// Mark the toEarlyStop tag here
diff --git a/src/nni_manager/yarn.lock b/src/nni_manager/yarn.lock
index deac0b5c89..379af7c4b9 100644
--- a/src/nni_manager/yarn.lock
+++ b/src/nni_manager/yarn.lock
@@ -157,6 +157,10 @@
version "7.0.3"
resolved "https://registry.yarnpkg.com/@types/json-schema/-/json-schema-7.0.3.tgz#bdfd69d61e464dcc81b25159c270d75a73c1a636"
+"@types/json5@^0.0.30":
+ version "0.0.30"
+ resolved "https://registry.yarnpkg.com/@types/json5/-/json5-0.0.30.tgz#44cb52f32a809734ca562e685c6473b5754a7818"
+
"@types/mime@*":
version "2.0.0"
resolved "https://registry.yarnpkg.com/@types/mime/-/mime-2.0.0.tgz#5a7306e367c539b9f6543499de8dd519fac37a8b"
@@ -1840,9 +1844,9 @@ growl@1.10.5:
version "1.10.5"
resolved "https://registry.yarnpkg.com/growl/-/growl-1.10.5.tgz#f2735dc2283674fa67478b10181059355c369e5e"
-handlebars@^4.0.11, handlebars@^4.3.0:
- version "4.5.3"
- resolved "https://registry.yarnpkg.com/handlebars/-/handlebars-4.5.3.tgz#5cf75bd8714f7605713511a56be7c349becb0482"
+handlebars@^4.0.11, handlebars@^4.5.3:
+ version "4.7.2"
+ resolved "https://registry.yarnpkg.com/handlebars/-/handlebars-4.7.2.tgz#01127b3840156a0927058779482031afe0e730d7"
dependencies:
neo-async "^2.6.0"
optimist "^0.6.1"
@@ -2371,6 +2375,12 @@ json-stringify-safe@~5.0.1:
version "5.0.1"
resolved "https://registry.yarnpkg.com/json-stringify-safe/-/json-stringify-safe-5.0.1.tgz#1296a2d58fd45f19a0f6ce01d65701e2c735b6eb"
+json5@^2.1.1:
+ version "2.1.1"
+ resolved "https://registry.yarnpkg.com/json5/-/json5-2.1.1.tgz#81b6cb04e9ba496f1c7005d07b4368a2638f90b6"
+ dependencies:
+ minimist "^1.2.0"
+
jsonparse@^1.2.0:
version "1.3.1"
resolved "https://registry.yarnpkg.com/jsonparse/-/jsonparse-1.3.1.tgz#3f4dae4a91fac315f71062f8521cc239f1366280"
diff --git a/src/sdk/pynni/nni/gridsearch_tuner/gridsearch_tuner.py b/src/sdk/pynni/nni/gridsearch_tuner/gridsearch_tuner.py
index f9dee6cdd8..8a9ab0a4ed 100644
--- a/src/sdk/pynni/nni/gridsearch_tuner/gridsearch_tuner.py
+++ b/src/sdk/pynni/nni/gridsearch_tuner/gridsearch_tuner.py
@@ -102,6 +102,9 @@ def _parse_randint(self, param_value):
"""
Parse type of randint parameter and return a list
"""
+ if param_value[0] >= param_value[1]:
+ raise ValueError("Randint should contain at least 1 candidate, but [%s, %s) contains none.",
+ param_value[0], param_value[1])
return np.arange(param_value[0], param_value[1]).tolist()
def _expand_parameters(self, para):
diff --git a/src/sdk/pynni/nni/hyperopt_tuner/hyperopt_tuner.py b/src/sdk/pynni/nni/hyperopt_tuner/hyperopt_tuner.py
index 466c29c484..c7e168191f 100644
--- a/src/sdk/pynni/nni/hyperopt_tuner/hyperopt_tuner.py
+++ b/src/sdk/pynni/nni/hyperopt_tuner/hyperopt_tuner.py
@@ -118,6 +118,8 @@ def json2vals(in_x, vals, out_y, name=NodeType.ROOT):
vals[NodeType.VALUE],
out_y,
name=name + '[%d]' % _index)
+ if _type == 'randint':
+ out_y[name] -= in_x[NodeType.VALUE][0]
else:
for key in in_x.keys():
json2vals(in_x[key], vals[key], out_y,
diff --git a/src/sdk/pynni/nni/msg_dispatcher.py b/src/sdk/pynni/nni/msg_dispatcher.py
index b3aadcaea1..739687fa0b 100644
--- a/src/sdk/pynni/nni/msg_dispatcher.py
+++ b/src/sdk/pynni/nni/msg_dispatcher.py
@@ -11,7 +11,7 @@
from .assessor import AssessResult
from .common import multi_thread_enabled, multi_phase_enabled
from .env_vars import dispatcher_env_vars
-from .utils import MetricType
+from .utils import MetricType, to_json
_logger = logging.getLogger(__name__)
@@ -62,7 +62,7 @@ def _pack_parameter(parameter_id, params, customized=False, trial_job_id=None, p
ret['parameter_index'] = parameter_index
else:
ret['parameter_index'] = 0
- return json_tricks.dumps(ret)
+ return to_json(ret)
class MsgDispatcher(MsgDispatcherBase):
diff --git a/src/sdk/pynni/nni/nas/pytorch/cdarts/__init__.py b/src/sdk/pynni/nni/nas/pytorch/cdarts/__init__.py
new file mode 100644
index 0000000000..2d00927846
--- /dev/null
+++ b/src/sdk/pynni/nni/nas/pytorch/cdarts/__init__.py
@@ -0,0 +1,5 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+from .mutator import RegularizedDartsMutator, RegularizedMutatorParallel, DartsDiscreteMutator
+from .trainer import CdartsTrainer
\ No newline at end of file
diff --git a/src/sdk/pynni/nni/nas/pytorch/cdarts/mutator.py b/src/sdk/pynni/nni/nas/pytorch/cdarts/mutator.py
new file mode 100644
index 0000000000..6010057828
--- /dev/null
+++ b/src/sdk/pynni/nni/nas/pytorch/cdarts/mutator.py
@@ -0,0 +1,146 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import torch
+
+from apex.parallel import DistributedDataParallel # pylint: disable=import-error
+from nni.nas.pytorch.darts import DartsMutator # pylint: disable=wrong-import-order
+from nni.nas.pytorch.mutables import LayerChoice # pylint: disable=wrong-import-order
+from nni.nas.pytorch.mutator import Mutator # pylint: disable=wrong-import-order
+
+
+class RegularizedDartsMutator(DartsMutator):
+ """
+ This is :class:`~nni.nas.pytorch.darts.DartsMutator` basically, with two differences.
+
+ 1. Choices can be cut (bypassed). This is done by ``cut_choices``. Cutted choices will not be used in
+ forward pass and thus consumes no memory.
+
+ 2. Regularization on choices, to prevent the mutator from overfitting on some choices.
+ """
+
+ def reset(self):
+ """
+ Warnings
+ --------
+ Renamed :func:`~reset_with_loss` to return regularization loss on reset.
+ """
+ raise ValueError("You should probably call `reset_with_loss`.")
+
+ def cut_choices(self, cut_num=2):
+ """
+ Cut the choices with the smallest weights.
+ ``cut_num`` should be the accumulative number of cutting, e.g., if first time cutting
+ is 2, the second time should be 4 to cut another two.
+
+ Parameters
+ ----------
+ cut_num : int
+ Number of choices to cut, so far.
+
+ Warnings
+ --------
+ Though the parameters are set to :math:`-\infty` to be bypassed, they will still receive gradient of 0,
+ which introduced ``nan`` problem when calling ``optimizer.step()``. To solve this issue, a simple way is to
+ reset nan to :math:`-\infty` each time after the parameters are updated.
+ """
+ # `cut_choices` is implemented but not used in current implementation of CdartsTrainer
+ for mutable in self.mutables:
+ if isinstance(mutable, LayerChoice):
+ _, idx = torch.topk(-self.choices[mutable.key], cut_num)
+ with torch.no_grad():
+ for i in idx:
+ self.choices[mutable.key][i] = -float("inf")
+
+ def reset_with_loss(self):
+ """
+ Resample and return loss. If loss is 0, to avoid device issue, it will return ``None``.
+
+ Currently loss penalty are proportional to the L1-norm of parameters corresponding
+ to modules if their type name contains certain substrings. These substrings include: ``poolwithoutbn``,
+ ``identity``, ``dilconv``.
+ """
+ self._cache, reg_loss = self.sample_search()
+ return reg_loss
+
+ def sample_search(self):
+ result = super().sample_search()
+ loss = []
+ for mutable in self.mutables:
+ if isinstance(mutable, LayerChoice):
+ def need_reg(choice):
+ return any(t in str(type(choice)).lower() for t in ["poolwithoutbn", "identity", "dilconv"])
+
+ for i, choice in enumerate(mutable.choices):
+ if need_reg(choice):
+ norm = torch.abs(self.choices[mutable.key][i])
+ if norm < 1E10:
+ loss.append(norm)
+ if not loss:
+ return result, None
+ return result, sum(loss)
+
+ def export(self, logger=None):
+ """
+ Export an architecture with logger. Genotype will be printed with logger.
+
+ Returns
+ -------
+ dict
+ A mapping from mutable keys to decisions.
+ """
+ result = self.sample_final()
+ if hasattr(self.model, "plot_genotype") and logger is not None:
+ genotypes = self.model.plot_genotype(result, logger)
+ return result, genotypes
+
+
+class RegularizedMutatorParallel(DistributedDataParallel):
+ """
+ Parallelize :class:`~RegularizedDartsMutator`.
+
+ This makes :func:`~RegularizedDartsMutator.reset_with_loss` method parallelized,
+ also allowing :func:`~RegularizedDartsMutator.cut_choices` and :func:`~RegularizedDartsMutator.export`
+ to be easily accessible.
+ """
+ def reset_with_loss(self):
+ """
+ Parallelized :func:`~RegularizedDartsMutator.reset_with_loss`.
+ """
+ result = self.module.reset_with_loss()
+ self.callback_queued = False
+ return result
+
+ def cut_choices(self, *args, **kwargs):
+ """
+ Parallelized :func:`~RegularizedDartsMutator.cut_choices`.
+ """
+ self.module.cut_choices(*args, **kwargs)
+
+ def export(self, logger):
+ """
+ Parallelized :func:`~RegularizedDartsMutator.export`.
+ """
+ return self.module.export(logger)
+
+
+class DartsDiscreteMutator(Mutator):
+ """
+ A mutator that applies the final sampling result of a parent mutator on another model to train.
+ """
+ def __init__(self, model, parent_mutator):
+ """
+ Initialization.
+
+ Parameters
+ ----------
+ model : nn.Module
+ The model to apply the mutator.
+ parent_mutator : Mutator
+ The mutator that provides ``sample_final`` method, that will be called to get the architecture.
+ """
+ super().__init__(model)
+ self.__dict__["parent_mutator"] = parent_mutator # avoid parameters to be included
+
+ def sample_search(self):
+ return self.parent_mutator.sample_final()
diff --git a/src/sdk/pynni/nni/nas/pytorch/cdarts/trainer.py b/src/sdk/pynni/nni/nas/pytorch/cdarts/trainer.py
new file mode 100644
index 0000000000..e050986b4c
--- /dev/null
+++ b/src/sdk/pynni/nni/nas/pytorch/cdarts/trainer.py
@@ -0,0 +1,275 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import json
+import logging
+import os
+
+import torch
+import torch.nn as nn
+import torch.nn.functional as F
+import apex # pylint: disable=import-error
+from apex.parallel import DistributedDataParallel # pylint: disable=import-error
+from nni.nas.pytorch.cdarts import RegularizedDartsMutator, RegularizedMutatorParallel, DartsDiscreteMutator # pylint: disable=wrong-import-order
+from nni.nas.pytorch.utils import AverageMeterGroup # pylint: disable=wrong-import-order
+
+from .utils import CyclicIterator, TorchTensorEncoder, accuracy, reduce_metrics
+
+PHASE_SMALL = "small"
+PHASE_LARGE = "large"
+
+
+class InteractiveKLLoss(nn.Module):
+ def __init__(self, temperature):
+ super().__init__()
+ self.temperature = temperature
+ # self.kl_loss = nn.KLDivLoss(reduction = 'batchmean')
+ self.kl_loss = nn.KLDivLoss()
+
+ def forward(self, student, teacher):
+ return self.kl_loss(F.log_softmax(student / self.temperature, dim=1),
+ F.softmax(teacher / self.temperature, dim=1))
+
+
+class CdartsTrainer(object):
+ def __init__(self, model_small, model_large, criterion, loaders, samplers, logger=None,
+ regular_coeff=5, regular_ratio=0.2, warmup_epochs=2, fix_head=True,
+ epochs=32, steps_per_epoch=None, loss_alpha=2, loss_T=2, distributed=True,
+ log_frequency=10, grad_clip=5.0, interactive_type='kl', output_path='./outputs',
+ w_lr=0.2, w_momentum=0.9, w_weight_decay=3e-4, alpha_lr=0.2, alpha_weight_decay=1e-4,
+ nasnet_lr=0.2, local_rank=0, share_module=True):
+ """
+ Initialize a CdartsTrainer.
+
+ Parameters
+ ----------
+ model_small : nn.Module
+ PyTorch model to be trained. This is the search network of CDARTS.
+ model_large : nn.Module
+ PyTorch model to be trained. This is the evaluation network of CDARTS.
+ criterion : callable
+ Receives logits and ground truth label, return a loss tensor, e.g., ``nn.CrossEntropyLoss()``.
+ loaders : list of torch.utils.data.DataLoader
+ List of train data and valid data loaders, for training weights and architecture weights respectively.
+ samplers : list of torch.utils.data.Sampler
+ List of train data and valid data samplers. This can be PyTorch standard samplers if not distributed.
+ In distributed mode, sampler needs to have ``set_epoch`` method. Refer to data utils in CDARTS example for details.
+ logger : logging.Logger
+ The logger for logging. Will use nni logger by default (if logger is ``None``).
+ regular_coeff : float
+ The coefficient of regular loss.
+ regular_ratio : float
+ The ratio of regular loss.
+ warmup_epochs : int
+ The epochs to warmup the search network
+ fix_head : bool
+ ``True`` if fixing the paramters of auxiliary heads, else unfix the paramters of auxiliary heads.
+ epochs : int
+ Number of epochs planned for training.
+ steps_per_epoch : int
+ Steps of one epoch.
+ loss_alpha : float
+ The loss coefficient.
+ loss_T : float
+ The loss coefficient.
+ distributed : bool
+ ``True`` if using distributed training, else non-distributed training.
+ log_frequency : int
+ Step count per logging.
+ grad_clip : float
+ Gradient clipping for weights.
+ interactive_type : string
+ ``kl`` or ``smoothl1``.
+ output_path : string
+ Log storage path.
+ w_lr : float
+ Learning rate of the search network parameters.
+ w_momentum : float
+ Momentum of the search and the evaluation network.
+ w_weight_decay : float
+ The weight decay the search and the evaluation network parameters.
+ alpha_lr : float
+ Learning rate of the architecture parameters.
+ alpha_weight_decay : float
+ The weight decay the architecture parameters.
+ nasnet_lr : float
+ Learning rate of the evaluation network parameters.
+ local_rank : int
+ The number of thread.
+ share_module : bool
+ ``True`` if sharing the stem and auxiliary heads, else not sharing these modules.
+ """
+ if logger is None:
+ logger = logging.getLogger(__name__)
+ train_loader, valid_loader = loaders
+ train_sampler, valid_sampler = samplers
+ self.train_loader = CyclicIterator(train_loader, train_sampler, distributed)
+ self.valid_loader = CyclicIterator(valid_loader, valid_sampler, distributed)
+
+ self.regular_coeff = regular_coeff
+ self.regular_ratio = regular_ratio
+ self.warmup_epochs = warmup_epochs
+ self.fix_head = fix_head
+ self.epochs = epochs
+ self.steps_per_epoch = steps_per_epoch
+ if self.steps_per_epoch is None:
+ self.steps_per_epoch = min(len(self.train_loader), len(self.valid_loader))
+ self.loss_alpha = loss_alpha
+ self.grad_clip = grad_clip
+ if interactive_type == "kl":
+ self.interactive_loss = InteractiveKLLoss(loss_T)
+ elif interactive_type == "smoothl1":
+ self.interactive_loss = nn.SmoothL1Loss()
+ self.loss_T = loss_T
+ self.distributed = distributed
+ self.log_frequency = log_frequency
+ self.main_proc = not distributed or local_rank == 0
+
+ self.logger = logger
+ self.checkpoint_dir = output_path
+ if self.main_proc:
+ os.makedirs(self.checkpoint_dir, exist_ok=True)
+ if distributed:
+ torch.distributed.barrier()
+
+ self.model_small = model_small
+ self.model_large = model_large
+ if self.fix_head:
+ for param in self.model_small.aux_head.parameters():
+ param.requires_grad = False
+ for param in self.model_large.aux_head.parameters():
+ param.requires_grad = False
+
+ self.mutator_small = RegularizedDartsMutator(self.model_small).cuda()
+ self.mutator_large = DartsDiscreteMutator(self.model_large, self.mutator_small).cuda()
+ self.criterion = criterion
+
+ self.optimizer_small = torch.optim.SGD(self.model_small.parameters(), w_lr,
+ momentum=w_momentum, weight_decay=w_weight_decay)
+ self.optimizer_large = torch.optim.SGD(self.model_large.parameters(), nasnet_lr,
+ momentum=w_momentum, weight_decay=w_weight_decay)
+ self.optimizer_alpha = torch.optim.Adam(self.mutator_small.parameters(), alpha_lr,
+ betas=(0.5, 0.999), weight_decay=alpha_weight_decay)
+
+ if distributed:
+ apex.parallel.convert_syncbn_model(self.model_small)
+ apex.parallel.convert_syncbn_model(self.model_large)
+ self.model_small = DistributedDataParallel(self.model_small, delay_allreduce=True)
+ self.model_large = DistributedDataParallel(self.model_large, delay_allreduce=True)
+ self.mutator_small = RegularizedMutatorParallel(self.mutator_small, delay_allreduce=True)
+ if share_module:
+ self.model_small.callback_queued = True
+ self.model_large.callback_queued = True
+ # mutator large never gets optimized, so do not need parallelized
+
+ def _warmup(self, phase, epoch):
+ assert phase in [PHASE_SMALL, PHASE_LARGE]
+ if phase == PHASE_SMALL:
+ model, optimizer = self.model_small, self.optimizer_small
+ elif phase == PHASE_LARGE:
+ model, optimizer = self.model_large, self.optimizer_large
+ model.train()
+ meters = AverageMeterGroup()
+ for step in range(self.steps_per_epoch):
+ x, y = next(self.train_loader)
+ x, y = x.cuda(), y.cuda()
+
+ optimizer.zero_grad()
+ logits_main, _ = model(x)
+ loss = self.criterion(logits_main, y)
+ loss.backward()
+
+ self._clip_grad_norm(model)
+ optimizer.step()
+ prec1, prec5 = accuracy(logits_main, y, topk=(1, 5))
+ metrics = {"prec1": prec1, "prec5": prec5, "loss": loss}
+ metrics = reduce_metrics(metrics, self.distributed)
+ meters.update(metrics)
+ if self.main_proc and (step % self.log_frequency == 0 or step + 1 == self.steps_per_epoch):
+ self.logger.info("Epoch [%d/%d] Step [%d/%d] (%s) %s", epoch + 1, self.epochs,
+ step + 1, self.steps_per_epoch, phase, meters)
+
+ def _clip_grad_norm(self, model):
+ if isinstance(model, DistributedDataParallel):
+ nn.utils.clip_grad_norm_(model.module.parameters(), self.grad_clip)
+ else:
+ nn.utils.clip_grad_norm_(model.parameters(), self.grad_clip)
+
+ def _reset_nan(self, parameters):
+ with torch.no_grad():
+ for param in parameters:
+ for i, p in enumerate(param):
+ if p != p: # equivalent to `isnan(p)`
+ param[i] = float("-inf")
+
+ def _joint_train(self, epoch):
+ self.model_large.train()
+ self.model_small.train()
+ meters = AverageMeterGroup()
+ for step in range(self.steps_per_epoch):
+ trn_x, trn_y = next(self.train_loader)
+ val_x, val_y = next(self.valid_loader)
+ trn_x, trn_y = trn_x.cuda(), trn_y.cuda()
+ val_x, val_y = val_x.cuda(), val_y.cuda()
+
+ # step 1. optimize architecture
+ self.optimizer_alpha.zero_grad()
+ self.optimizer_large.zero_grad()
+ reg_decay = max(self.regular_coeff * (1 - float(epoch - self.warmup_epochs) / (
+ (self.epochs - self.warmup_epochs) * self.regular_ratio)), 0)
+ loss_regular = self.mutator_small.reset_with_loss()
+ if loss_regular:
+ loss_regular *= reg_decay
+ logits_search, emsemble_logits_search = self.model_small(val_x)
+ logits_main, emsemble_logits_main = self.model_large(val_x)
+ loss_cls = (self.criterion(logits_search, val_y) + self.criterion(logits_main, val_y)) / self.loss_alpha
+ loss_interactive = self.interactive_loss(emsemble_logits_search, emsemble_logits_main) * (self.loss_T ** 2) * self.loss_alpha
+ loss = loss_cls + loss_interactive + loss_regular
+ loss.backward()
+ self._clip_grad_norm(self.model_large)
+ self.optimizer_large.step()
+ self.optimizer_alpha.step()
+ # NOTE: need to call here `self._reset_nan(self.mutator_small.parameters())` if `cut_choices`
+
+ # step 2. optimize op weights
+ self.optimizer_small.zero_grad()
+ with torch.no_grad():
+ # resample architecture since parameters have been changed
+ self.mutator_small.reset_with_loss()
+ logits_search_train, _ = self.model_small(trn_x)
+ loss_weight = self.criterion(logits_search_train, trn_y)
+ loss_weight.backward()
+ self._clip_grad_norm(self.model_small)
+ self.optimizer_small.step()
+
+ metrics = {"loss_cls": loss_cls, "loss_interactive": loss_interactive,
+ "loss_regular": loss_regular, "loss_weight": loss_weight}
+ metrics = reduce_metrics(metrics, self.distributed)
+ meters.update(metrics)
+
+ if self.main_proc and (step % self.log_frequency == 0 or step + 1 == self.steps_per_epoch):
+ self.logger.info("Epoch [%d/%d] Step [%d/%d] (joint) %s", epoch + 1, self.epochs,
+ step + 1, self.steps_per_epoch, meters)
+
+ def train(self):
+ for epoch in range(self.epochs):
+ if epoch < self.warmup_epochs:
+ with torch.no_grad(): # otherwise grads will be retained on the architecture params
+ self.mutator_small.reset_with_loss()
+ self._warmup(PHASE_SMALL, epoch)
+ else:
+ with torch.no_grad():
+ self.mutator_large.reset()
+ self._warmup(PHASE_LARGE, epoch)
+ self._joint_train(epoch)
+
+ self.export(os.path.join(self.checkpoint_dir, "epoch_{:02d}.json".format(epoch)),
+ os.path.join(self.checkpoint_dir, "epoch_{:02d}.genotypes".format(epoch)))
+
+ def export(self, file, genotype_file):
+ if self.main_proc:
+ mutator_export, genotypes = self.mutator_small.export(self.logger)
+ with open(file, "w") as f:
+ json.dump(mutator_export, f, indent=2, sort_keys=True, cls=TorchTensorEncoder)
+ with open(genotype_file, "w") as f:
+ f.write(str(genotypes))
diff --git a/src/sdk/pynni/nni/nas/pytorch/cdarts/utils.py b/src/sdk/pynni/nni/nas/pytorch/cdarts/utils.py
new file mode 100644
index 0000000000..780f6fdc0e
--- /dev/null
+++ b/src/sdk/pynni/nni/nas/pytorch/cdarts/utils.py
@@ -0,0 +1,76 @@
+# Copyright (c) Microsoft Corporation.
+# Licensed under the MIT license.
+
+import json
+import os
+
+import torch
+import torch.distributed as dist
+
+
+class CyclicIterator:
+ def __init__(self, loader, sampler, distributed):
+ self.loader = loader
+ self.sampler = sampler
+ self.epoch = 0
+ self.distributed = distributed
+ self._next_epoch()
+
+ def _next_epoch(self):
+ if self.distributed:
+ self.sampler.set_epoch(self.epoch)
+ self.iterator = iter(self.loader)
+ self.epoch += 1
+
+ def __len__(self):
+ return len(self.loader)
+
+ def __iter__(self):
+ return self
+
+ def __next__(self):
+ try:
+ return next(self.iterator)
+ except StopIteration:
+ self._next_epoch()
+ return next(self.iterator)
+
+
+class TorchTensorEncoder(json.JSONEncoder):
+ def default(self, o): # pylint: disable=method-hidden
+ if isinstance(o, torch.Tensor):
+ return o.tolist()
+ return super().default(o)
+
+
+def accuracy(output, target, topk=(1,)):
+ """ Computes the precision@k for the specified values of k """
+ maxk = max(topk)
+ batch_size = target.size(0)
+
+ _, pred = output.topk(maxk, 1, True, True)
+ pred = pred.t()
+ # one-hot case
+ if target.ndimension() > 1:
+ target = target.max(1)[1]
+
+ correct = pred.eq(target.view(1, -1).expand_as(pred))
+
+ res = []
+ for k in topk:
+ correct_k = correct[:k].view(-1).float().sum(0)
+ res.append(correct_k.mul_(1.0 / batch_size))
+ return res
+
+
+def reduce_tensor(tensor):
+ rt = tensor.clone()
+ dist.all_reduce(rt, op=dist.ReduceOp.SUM)
+ rt /= float(os.environ["WORLD_SIZE"])
+ return rt
+
+
+def reduce_metrics(metrics, distributed=False):
+ if distributed:
+ return {k: reduce_tensor(v).item() for k, v in metrics.items()}
+ return {k: v.item() for k, v in metrics.items()}
diff --git a/src/sdk/pynni/nni/platform/local.py b/src/sdk/pynni/nni/platform/local.py
index 556ff1398d..1b4bc081c5 100644
--- a/src/sdk/pynni/nni/platform/local.py
+++ b/src/sdk/pynni/nni/platform/local.py
@@ -6,10 +6,10 @@
import json
import time
import subprocess
-import json_tricks
from ..common import init_logger
from ..env_vars import trial_env_vars
+from ..utils import to_json
_sysdir = trial_env_vars.NNI_SYS_DIR
if not os.path.exists(os.path.join(_sysdir, '.nni')):
@@ -30,7 +30,7 @@
_param_index = 0
def request_next_parameter():
- metric = json_tricks.dumps({
+ metric = to_json({
'trial_job_id': trial_env_vars.NNI_TRIAL_JOB_ID,
'type': 'REQUEST_PARAMETER',
'sequence': 0,
diff --git a/src/sdk/pynni/nni/trial.py b/src/sdk/pynni/nni/trial.py
index 6feed9dbef..70127330fa 100644
--- a/src/sdk/pynni/nni/trial.py
+++ b/src/sdk/pynni/nni/trial.py
@@ -1,8 +1,7 @@
# Copyright (c) Microsoft Corporation.
# Licensed under the MIT license.
-import json_tricks
-
+from .utils import to_json
from .env_vars import trial_env_vars
from . import platform
@@ -110,7 +109,7 @@ def report_intermediate_result(metric):
global _intermediate_seq
assert _params or trial_env_vars.NNI_PLATFORM is None, \
'nni.get_next_parameter() needs to be called before report_intermediate_result'
- metric = json_tricks.dumps({
+ metric = to_json({
'parameter_id': _params['parameter_id'] if _params else None,
'trial_job_id': trial_env_vars.NNI_TRIAL_JOB_ID,
'type': 'PERIODICAL',
@@ -120,7 +119,6 @@ def report_intermediate_result(metric):
_intermediate_seq += 1
platform.send_metric(metric)
-
def report_final_result(metric):
"""
Reports final result to NNI.
@@ -132,7 +130,7 @@ def report_final_result(metric):
"""
assert _params or trial_env_vars.NNI_PLATFORM is None, \
'nni.get_next_parameter() needs to be called before report_final_result'
- metric = json_tricks.dumps({
+ metric = to_json({
'parameter_id': _params['parameter_id'] if _params else None,
'trial_job_id': trial_env_vars.NNI_TRIAL_JOB_ID,
'type': 'FINAL',
diff --git a/src/sdk/pynni/nni/utils.py b/src/sdk/pynni/nni/utils.py
index 99b5017107..bb7602a4b1 100644
--- a/src/sdk/pynni/nni/utils.py
+++ b/src/sdk/pynni/nni/utils.py
@@ -1,16 +1,16 @@
# Copyright (c) Microsoft Corporation.
# Licensed under the MIT license.
-"""
-utils.py
-"""
-
import os
+import functools
from enum import Enum, unique
+import json_tricks
from .common import init_logger
from .env_vars import dispatcher_env_vars
+to_json = functools.partial(json_tricks.dumps, allow_nan=True)
+
@unique
class OptimizeMode(Enum):
"""Optimize Mode class
diff --git a/src/sdk/pynni/tests/assets/search_space.json b/src/sdk/pynni/tests/assets/search_space.json
index 0e7c7ba9cc..21b6f90996 100644
--- a/src/sdk/pynni/tests/assets/search_space.json
+++ b/src/sdk/pynni/tests/assets/search_space.json
@@ -1,8 +1,7 @@
{
"choice_str": {
"_type": "choice",
- "_value": ["cat", "dog", "elephant", "cow", "sheep", "panda"],
- "fail": ["metis", "gp"]
+ "_value": ["cat", "dog", "elephant", "cow", "sheep", "panda"]
},
"choice_int": {
"_type": "choice",
@@ -10,8 +9,7 @@
},
"choice_mixed": {
"_type": "choice",
- "_value": [0.3, "cat", 1, null],
- "fail": ["metis", "gp"]
+ "_value": [0.3, "cat", 1, null]
},
"choice_float": {
"_type": "choice",
diff --git a/src/sdk/pynni/tests/test_builtin_tuners.py b/src/sdk/pynni/tests/test_builtin_tuners.py
index 8dd3731983..2fe2c8345c 100644
--- a/src/sdk/pynni/tests/test_builtin_tuners.py
+++ b/src/sdk/pynni/tests/test_builtin_tuners.py
@@ -5,6 +5,7 @@
import json
import logging
import os
+import random
import shutil
import sys
from unittest import TestCase, main
@@ -15,6 +16,7 @@
from nni.gridsearch_tuner.gridsearch_tuner import GridSearchTuner
from nni.hyperopt_tuner.hyperopt_tuner import HyperoptTuner
from nni.metis_tuner.metis_tuner import MetisTuner
+
try:
from nni.smac_tuner.smac_tuner import SMACTuner
except ImportError:
@@ -34,20 +36,28 @@ class BuiltinTunersTestCase(TestCase):
- [X] generate_multiple_parameters
- [ ] import_data
- [ ] trial_end
- - [ ] receive_trial_result
+ - [x] receive_trial_result
"""
+ def setUp(self):
+ self.test_round = 3
+ self.params_each_round = 50
+ self.exhaustive = False
+
def search_space_test_one(self, tuner_factory, search_space):
tuner = tuner_factory()
self.assertIsInstance(tuner, Tuner)
tuner.update_search_space(search_space)
- parameters = tuner.generate_multiple_parameters(list(range(0, 50)))
- logger.info(parameters)
- self.check_range(parameters, search_space)
- if not parameters: # TODO: not strict
- raise ValueError("No parameters generated")
- return parameters
+ for i in range(self.test_round):
+ parameters = tuner.generate_multiple_parameters(list(range(i * self.params_each_round,
+ (i + 1) * self.params_each_round)))
+ logger.debug(parameters)
+ self.check_range(parameters, search_space)
+ for k in range(min(len(parameters), self.params_each_round)):
+ tuner.receive_trial_result(self.params_each_round * i + k, parameters[k], random.uniform(-100, 100))
+ if not parameters and not self.exhaustive:
+ raise ValueError("No parameters generated")
def check_range(self, generated_params, search_space):
EPS = 1E-6
@@ -91,7 +101,8 @@ def check_range(self, generated_params, search_space):
for layer_name in item["_value"].keys():
self.assertIn(v[layer_name]["chosen_layer"], item["layer_choice"])
- def search_space_test_all(self, tuner_factory, supported_types=None, ignore_types=None):
+ def search_space_test_all(self, tuner_factory, supported_types=None, ignore_types=None, fail_types=None):
+ # Three types: 1. supported; 2. ignore; 3. fail.
# NOTE(yuge): ignore types
# Supported types are listed in the table. They are meant to be supported and should be correct.
# Other than those, all the rest are "unsupported", which are expected to produce ridiculous results
@@ -103,16 +114,18 @@ def search_space_test_all(self, tuner_factory, supported_types=None, ignore_type
if supported_types is None:
supported_types = ["choice", "randint", "uniform", "quniform", "loguniform", "qloguniform",
"normal", "qnormal", "lognormal", "qlognormal"]
+ if fail_types is None:
+ fail_types = []
+ if ignore_types is None:
+ ignore_types = []
full_supported_search_space = dict()
for single in search_space_all:
- single_keyword = single.split("_")
space = search_space_all[single]
- expected_fail = not any([t in single_keyword for t in supported_types]) or "fail" in single_keyword
- if ignore_types is not None and any([t in ignore_types for t in single_keyword]):
+ if any(single.startswith(t) for t in ignore_types):
continue
- if "fail" in space:
- if self._testMethodName.split("_", 1)[1] in space.pop("fail"):
- expected_fail = True
+ expected_fail = not any(single.startswith(t) for t in supported_types) or \
+ any(single.startswith(t) for t in fail_types) or \
+ "fail" in single # name contains fail (fail on all)
single_search_space = {single: space}
if not expected_fail:
# supports this key
@@ -129,11 +142,14 @@ def search_space_test_all(self, tuner_factory, supported_types=None, ignore_type
self.search_space_test_one(tuner_factory, full_supported_search_space)
def test_grid_search(self):
+ self.exhaustive = True
self.search_space_test_all(lambda: GridSearchTuner(),
supported_types=["choice", "randint", "quniform"])
def test_tpe(self):
- self.search_space_test_all(lambda: HyperoptTuner("tpe"))
+ self.search_space_test_all(lambda: HyperoptTuner("tpe"),
+ ignore_types=["uniform_equal", "qloguniform_equal", "loguniform_equal", "quniform_clip_2"])
+ # NOTE: types are ignored because `tpe.py line 465, in adaptive_parzen_normal assert prior_sigma > 0`
def test_random_search(self):
self.search_space_test_all(lambda: HyperoptTuner("random_search"))
@@ -148,6 +164,7 @@ def test_smac(self):
supported_types=["choice", "randint", "uniform", "quniform", "loguniform"])
def test_batch(self):
+ self.exhaustive = True
self.search_space_test_all(lambda: BatchTuner(),
supported_types=["choice"])
@@ -156,14 +173,18 @@ def test_evolution(self):
self.search_space_test_all(lambda: EvolutionTuner(population_size=100))
def test_gp(self):
+ self.test_round = 1 # NOTE: GP tuner got hanged for multiple testing round
self.search_space_test_all(lambda: GPTuner(),
supported_types=["choice", "randint", "uniform", "quniform", "loguniform",
"qloguniform"],
- ignore_types=["normal", "lognormal", "qnormal", "qlognormal"])
+ ignore_types=["normal", "lognormal", "qnormal", "qlognormal"],
+ fail_types=["choice_str", "choice_mixed"])
def test_metis(self):
+ self.test_round = 1 # NOTE: Metis tuner got hanged for multiple testing round
self.search_space_test_all(lambda: MetisTuner(),
- supported_types=["choice", "randint", "uniform", "quniform"])
+ supported_types=["choice", "randint", "uniform", "quniform"],
+ fail_types=["choice_str", "choice_mixed"])
def test_networkmorphism(self):
pass
diff --git a/src/webui/package.json b/src/webui/package.json
index 955b0ab4f2..ad202d89f3 100644
--- a/src/webui/package.json
+++ b/src/webui/package.json
@@ -18,6 +18,7 @@
"fork-ts-checker-webpack-plugin": "^1.5.0",
"fs-extra": "^8.1.0",
"html-webpack-plugin": "^4.0.0-beta.8",
+ "json5": "^2.1.1",
"less": "^3.9.0",
"less-loader": "^5.0.0",
"mini-css-extract-plugin": "^0.8.0",
@@ -55,6 +56,7 @@
"eslint": "npx eslint ./ --ext .tsx,.ts"
},
"devDependencies": {
+ "@types/json5": "^0.0.30",
"@types/node": "^10.14.14",
"@types/react": "16.4.17",
"@types/react-dom": "^16.0.7",
diff --git a/src/webui/src/components/trial-detail/TableList.tsx b/src/webui/src/components/trial-detail/TableList.tsx
index 757da666fa..2b98b2b894 100644
--- a/src/webui/src/components/trial-detail/TableList.tsx
+++ b/src/webui/src/components/trial-detail/TableList.tsx
@@ -6,7 +6,7 @@ import { ColumnProps } from 'antd/lib/table';
const Option = Select.Option;
const CheckboxGroup = Checkbox.Group;
import { MANAGER_IP, trialJobStatus, COLUMN_INDEX, COLUMNPro } from '../../static/const';
-import { convertDuration, formatTimestamp, intermediateGraphOption, killJob } from '../../static/function';
+import { convertDuration, formatTimestamp, intermediateGraphOption, killJob, parseMetrics } from '../../static/function';
import { EXPERIMENT, TRIALS } from '../../static/datamodel';
import { TableRecord } from '../../static/interface';
import OpenRow from '../public-child/OpenRow';
@@ -178,11 +178,11 @@ class TableList extends React.Component {
// get intermediate result dict keys array
let otherkeys: Array = ['default'];
if (res.data.length !== 0) {
- otherkeys = Object.keys(JSON.parse(res.data[0].data));
+ otherkeys = Object.keys(parseMetrics(res.data[0].data));
}
// intermediateArr just store default val
Object.keys(res.data).map(item => {
- const temp = JSON.parse(res.data[item].data);
+ const temp = parseMetrics(res.data[item].data);
if (typeof temp === 'object') {
intermediateArr.push(temp.default);
} else {
@@ -210,7 +210,7 @@ class TableList extends React.Component {
// just watch default key-val
if (isShowDefault === true) {
Object.keys(intermediateData).map(item => {
- const temp = JSON.parse(intermediateData[item].data);
+ const temp = parseMetrics(intermediateData[item].data);
if (typeof temp === 'object') {
intermediateArr.push(temp[value]);
} else {
@@ -219,7 +219,7 @@ class TableList extends React.Component {
});
} else {
Object.keys(intermediateData).map(item => {
- const temp = JSON.parse(intermediateData[item].data);
+ const temp = parseMetrics(intermediateData[item].data);
if (typeof temp === 'object') {
intermediateArr.push(temp[value]);
}
@@ -431,7 +431,8 @@ class TableList extends React.Component {
key: 'operation',
render: (text: string, record: TableRecord) => {
const trialStatus = record.status;
- const flag: boolean = (trialStatus === 'RUNNING') ? false : true;
+ // could kill a job when its status is RUNNING or UNKNOWN
+ const flag: boolean = (trialStatus === 'RUNNING' || trialStatus === 'UNKNOWN') ? false : true;
return (
{/* see intermediate result graph */}
diff --git a/src/webui/src/static/function.ts b/src/webui/src/static/function.ts
index fd1ccff682..33193f2bf4 100644
--- a/src/webui/src/static/function.ts
+++ b/src/webui/src/static/function.ts
@@ -1,3 +1,4 @@
+import * as JSON5 from 'json5';
import axios from 'axios';
import { message } from 'antd';
import { MANAGER_IP } from './const';
@@ -173,8 +174,16 @@ function formatTimestamp(timestamp?: number, placeholder?: string = 'N/A'): stri
return timestamp ? new Date(timestamp).toLocaleString('en-US') : placeholder;
}
+function parseMetrics(metricData: string): any {
+ if (metricData.includes('NaN')) {
+ return JSON5.parse(metricData)
+ } else {
+ return JSON.parse(metricData)
+ }
+}
+
function metricAccuracy(metric: MetricDataRecord): number {
- const data = JSON.parse(metric.data);
+ const data = parseMetrics(metric.data);
return typeof data === 'number' ? data : NaN;
}
@@ -186,5 +195,5 @@ function formatAccuracy(accuracy: number): string {
export {
convertTime, convertDuration, getFinalResult, getFinal, downFile,
intermediateGraphOption, killJob, filterByStatus, filterDuration,
- formatAccuracy, formatTimestamp, metricAccuracy
+ formatAccuracy, formatTimestamp, metricAccuracy, parseMetrics
};
diff --git a/src/webui/src/static/model/trial.ts b/src/webui/src/static/model/trial.ts
index 2a91450ebc..2ab0e4afa8 100644
--- a/src/webui/src/static/model/trial.ts
+++ b/src/webui/src/static/model/trial.ts
@@ -1,5 +1,5 @@
import { MetricDataRecord, TrialJobInfo, TableObj, TableRecord, Parameters, FinalType } from '../interface';
-import { getFinal, formatAccuracy, metricAccuracy } from '../function';
+import { getFinal, formatAccuracy, metricAccuracy, parseMetrics } from '../function';
class Trial implements TableObj {
private metricsInitialized: boolean = false;
@@ -56,7 +56,7 @@ class Trial implements TableObj {
// TODO: support intermeidate result is dict
const temp = this.intermediates[this.intermediates.length - 1];
if (temp !== undefined) {
- return JSON.parse(temp.data);
+ return parseMetrics(temp.data);
} else {
return undefined;
}
@@ -138,10 +138,10 @@ class Trial implements TableObj {
const mediate: number[] = [ ];
for (const items of this.intermediateMetrics) {
- if (typeof JSON.parse(items.data) === 'object') {
- mediate.push(JSON.parse(items.data).default);
+ if (typeof parseMetrics(items.data) === 'object') {
+ mediate.push(parseMetrics(items.data).default);
} else {
- mediate.push(JSON.parse(items.data));
+ mediate.push(parseMetrics(items.data));
}
}
ret.intermediate = mediate;
diff --git a/src/webui/yarn.lock b/src/webui/yarn.lock
index 0c8fbd6672..1d7548df58 100644
--- a/src/webui/yarn.lock
+++ b/src/webui/yarn.lock
@@ -80,6 +80,10 @@
version "0.0.29"
resolved "https://registry.yarnpkg.com/@types/json5/-/json5-0.0.29.tgz#ee28707ae94e11d2b827bcbe5270bcea7f3e71ee"
+"@types/json5@^0.0.30":
+ version "0.0.30"
+ resolved "https://registry.yarnpkg.com/@types/json5/-/json5-0.0.30.tgz#44cb52f32a809734ca562e685c6473b5754a7818"
+
"@types/minimatch@*":
version "3.0.3"
resolved "https://registry.yarnpkg.com/@types/minimatch/-/minimatch-3.0.3.tgz#3dca0e3f33b200fc7d1139c0cd96c1268cadfd9d"
@@ -3988,6 +3992,12 @@ json5@^1.0.1:
dependencies:
minimist "^1.2.0"
+json5@^2.1.1:
+ version "2.1.1"
+ resolved "https://registry.yarnpkg.com/json5/-/json5-2.1.1.tgz#81b6cb04e9ba496f1c7005d07b4368a2638f90b6"
+ dependencies:
+ minimist "^1.2.0"
+
jsonfile@^4.0.0:
version "4.0.0"
resolved "https://registry.yarnpkg.com/jsonfile/-/jsonfile-4.0.0.tgz#8771aae0799b64076b76640fca058f9c10e33ecb"
diff --git a/tools/nni_cmd/config_schema.py b/tools/nni_cmd/config_schema.py
index 8017946ce9..30db148200 100644
--- a/tools/nni_cmd/config_schema.py
+++ b/tools/nni_cmd/config_schema.py
@@ -407,15 +407,8 @@ def setPathCheck(key):
}
machine_list_schema = {
- Optional('machineList'):[Or({
- 'ip': setType('ip', str),
- Optional('port'): setNumberRange('port', int, 1, 65535),
- 'username': setType('username', str),
- 'passwd': setType('passwd', str),
- Optional('gpuIndices'): Or(int, And(str, lambda x: len([int(i) for i in x.split(',')]) > 0), error='gpuIndex format error!'),
- Optional('maxTrialNumPerGpu'): setType('maxTrialNumPerGpu', int),
- Optional('useActiveGpu'): setType('useActiveGpu', bool)
- }, {
+ Optional('machineList'):[Or(
+ {
'ip': setType('ip', str),
Optional('port'): setNumberRange('port', int, 1, 65535),
'username': setType('username', str),
@@ -424,6 +417,15 @@ def setPathCheck(key):
Optional('gpuIndices'): Or(int, And(str, lambda x: len([int(i) for i in x.split(',')]) > 0), error='gpuIndex format error!'),
Optional('maxTrialNumPerGpu'): setType('maxTrialNumPerGpu', int),
Optional('useActiveGpu'): setType('useActiveGpu', bool)
+ },
+ {
+ 'ip': setType('ip', str),
+ Optional('port'): setNumberRange('port', int, 1, 65535),
+ 'username': setType('username', str),
+ 'passwd': setType('passwd', str),
+ Optional('gpuIndices'): Or(int, And(str, lambda x: len([int(i) for i in x.split(',')]) > 0), error='gpuIndex format error!'),
+ Optional('maxTrialNumPerGpu'): setType('maxTrialNumPerGpu', int),
+ Optional('useActiveGpu'): setType('useActiveGpu', bool)
})]
}
diff --git a/tools/nni_cmd/config_utils.py b/tools/nni_cmd/config_utils.py
index 1b7cbc1662..8cc1dc8ada 100644
--- a/tools/nni_cmd/config_utils.py
+++ b/tools/nni_cmd/config_utils.py
@@ -76,7 +76,7 @@ def update_experiment(self, expId, key, value):
def remove_experiment(self, expId):
'''remove an experiment by id'''
- if id in self.experiments:
+ if expId in self.experiments:
self.experiments.pop(expId)
self.write_file()
diff --git a/tools/nni_cmd/nnictl_utils.py b/tools/nni_cmd/nnictl_utils.py
index a66197fac9..4866bcdce4 100644
--- a/tools/nni_cmd/nnictl_utils.py
+++ b/tools/nni_cmd/nnictl_utils.py
@@ -403,11 +403,13 @@ def remote_clean(machine_list, experiment_id=None):
userName = machine.get('username')
host = machine.get('ip')
port = machine.get('port')
+ sshKeyPath = machine.get('sshKeyPath')
+ passphrase = machine.get('passphrase')
if experiment_id:
remote_dir = '/' + '/'.join(['tmp', 'nni', 'experiments', experiment_id])
else:
remote_dir = '/' + '/'.join(['tmp', 'nni', 'experiments'])
- sftp = create_ssh_sftp_client(host, port, userName, passwd)
+ sftp = create_ssh_sftp_client(host, port, userName, passwd, sshKeyPath, passphrase)
print_normal('removing folder {0}'.format(host + ':' + str(port) + remote_dir))
remove_remote_directory(sftp, remote_dir)
diff --git a/tools/nni_cmd/ssh_utils.py b/tools/nni_cmd/ssh_utils.py
index 2e68611206..e3f26a8e24 100644
--- a/tools/nni_cmd/ssh_utils.py
+++ b/tools/nni_cmd/ssh_utils.py
@@ -30,12 +30,16 @@ def copy_remote_directory_to_local(sftp, remote_path, local_path):
except Exception:
pass
-def create_ssh_sftp_client(host_ip, port, username, password):
+def create_ssh_sftp_client(host_ip, port, username, password, ssh_key_path, passphrase):
'''create ssh client'''
try:
paramiko = check_environment()
conn = paramiko.Transport(host_ip, port)
- conn.connect(username=username, password=password)
+ if ssh_key_path is not None:
+ ssh_key = paramiko.RSAKey.from_private_key_file(ssh_key_path, password=passphrase)
+ conn.connect(username=username, pkey=ssh_key)
+ else:
+ conn.connect(username=username, password=password)
sftp = paramiko.SFTPClient.from_transport(conn)
return sftp
except Exception as exception:
diff --git a/tools/nni_cmd/tensorboard_utils.py b/tools/nni_cmd/tensorboard_utils.py
index 8cb0bbfc17..60d589083a 100644
--- a/tools/nni_cmd/tensorboard_utils.py
+++ b/tools/nni_cmd/tensorboard_utils.py
@@ -37,12 +37,14 @@ def copy_data_from_remote(args, nni_config, trial_content, path_list, host_list,
machine_dict = {}
local_path_list = []
for machine in machine_list:
- machine_dict[machine['ip']] = {'port': machine['port'], 'passwd': machine['passwd'], 'username': machine['username']}
+ machine_dict[machine['ip']] = {'port': machine['port'], 'passwd': machine['passwd'], 'username': machine['username'],
+ 'sshKeyPath': machine.get('sshKeyPath'), 'passphrase': machine.get('passphrase')}
for index, host in enumerate(host_list):
local_path = os.path.join(temp_nni_path, trial_content[index].get('id'))
local_path_list.append(local_path)
print_normal('Copying log data from %s to %s' % (host + ':' + path_list[index], local_path))
- sftp = create_ssh_sftp_client(host, machine_dict[host]['port'], machine_dict[host]['username'], machine_dict[host]['passwd'])
+ sftp = create_ssh_sftp_client(host, machine_dict[host]['port'], machine_dict[host]['username'], machine_dict[host]['passwd'],
+ machine_dict[host]['sshKeyPath'], machine_dict[host]['passphrase'])
copy_remote_directory_to_local(sftp, path_list[index], local_path)
print_normal('Copy done!')
return local_path_list