-
-
Notifications
You must be signed in to change notification settings - Fork 1.1k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
N-dimensional boolean indexing #5179
Comments
|
@max-sixty The reason is that my method is basically a special case of point-wise indexing: core_dim_locs = {key: value for key, value in core_dim_locs_from_cond(mask, new_dim_name="newdim")}
# pointwise selection
data.sel(
dim_0=outliers_subset["dim_0"],
dim_1=outliers_subset["dim_1"],
dim_2=outliers_subset["dim_2"]
)(Note that you loose chunk information by this method, that's why it is less efficient) When you want to select random items from a N-dimensional array, you can either model the result as some sparse array or by stacking the dimensions. |
|
Ah right, I see now, thanks for explaining. Allowing pointwise indexing with bool indexes would also be welcome. |
|
I wonder if this is just a better proposal than making N-dimensional boolean indexing an alias for |
|
@max-sixty and I have been having some more discussion about whether this is what But regardless of what we want boolean indexing with |
|
fyi, I updated the boolean indexing to support additional or missing dimensions: |
Currently, the docs state that boolean indexing is only possible with 1-dimensional arrays:
http://xarray.pydata.org/en/stable/indexing.html
However, I often have the case where I'd like to convert a subset of an xarray to a dataframe.
Usually, I would call e.g.:
However, this approach is incredibly slow and memory-demanding, since it creates a MultiIndex of every possible coordinate in the array.
Describe the solution you'd like
A better approach would be to directly allow index selection with the boolean array:
This way, it is possible to
np.argwhere()variable.data[mask]Additional context
I created a proof-of-concept that works for my projects:
https://gist.github.com/Hoeze/c746ea1e5fef40d99997f765c48d3c0d
Some important lines are those:
As a result, I would expect something like this:
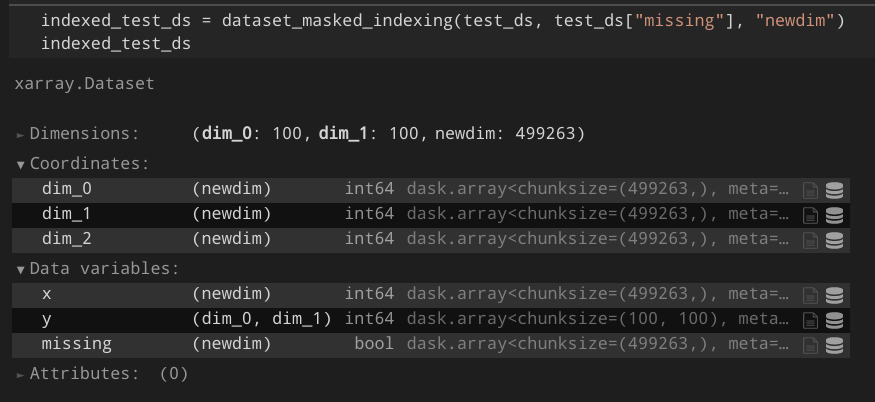
The text was updated successfully, but these errors were encountered: