diff --git a/.github/workflows/ci-tests.yml b/.github/workflows/ci-tests.yml
index cee98dc33d..81f5132ccf 100644
--- a/.github/workflows/ci-tests.yml
+++ b/.github/workflows/ci-tests.yml
@@ -36,8 +36,12 @@ jobs:
matrix:
os: ["ubuntu-latest"]
python-version: ["3.10"]
- session: ["tests", "doctest", "gallery", "linkcheck"]
+ session: ["doctest", "gallery", "linkcheck"]
include:
+ - os: "ubuntu-latest"
+ python-version: "3.10"
+ session: "tests"
+ coverage: "--coverage"
- os: "ubuntu-latest"
python-version: "3.9"
session: "tests"
@@ -133,4 +137,8 @@ jobs:
env:
PY_VER: ${{ matrix.python-version }}
run: |
- nox --session ${{ matrix.session }} -- --verbose
+ nox --session ${{ matrix.session }} -- --verbose ${{ matrix.coverage }}
+
+ - name: Upload coverage report
+ uses: codecov/codecov-action@v3
+ if: ${{ matrix.coverage }}
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
index ad5a9d4626..7c95eeaca3 100644
--- a/.pre-commit-config.yaml
+++ b/.pre-commit-config.yaml
@@ -29,7 +29,7 @@ repos:
- id: no-commit-to-branch
- repo: https://github.com/psf/black
- rev: 22.12.0
+ rev: 23.1.0
hooks:
- id: black
pass_filenames: false
@@ -43,18 +43,17 @@ repos:
args: [--config=./setup.cfg]
- repo: https://github.com/pycqa/isort
- rev: 5.11.4
+ rev: 5.12.0
hooks:
- id: isort
types: [file, python]
args: [--filter-files]
- repo: https://github.com/asottile/blacken-docs
- rev: v1.12.1
+ rev: 1.13.0
hooks:
- id: blacken-docs
types: [file, rst]
- additional_dependencies: [black==21.6b0]
- repo: https://github.com/aio-libs/sort-all
rev: v1.2.0
diff --git a/README.md b/README.md
index ac2781f469..67c4399116 100644
--- a/README.md
+++ b/README.md
@@ -19,6 +19,9 @@
 +
+
+
+
@@ -54,3 +57,24 @@ For documentation see the
developer version or the most recent released
stable version.
+
+## [#ShowYourStripes](https://showyourstripes.info/s/globe)
+
+
+
+ 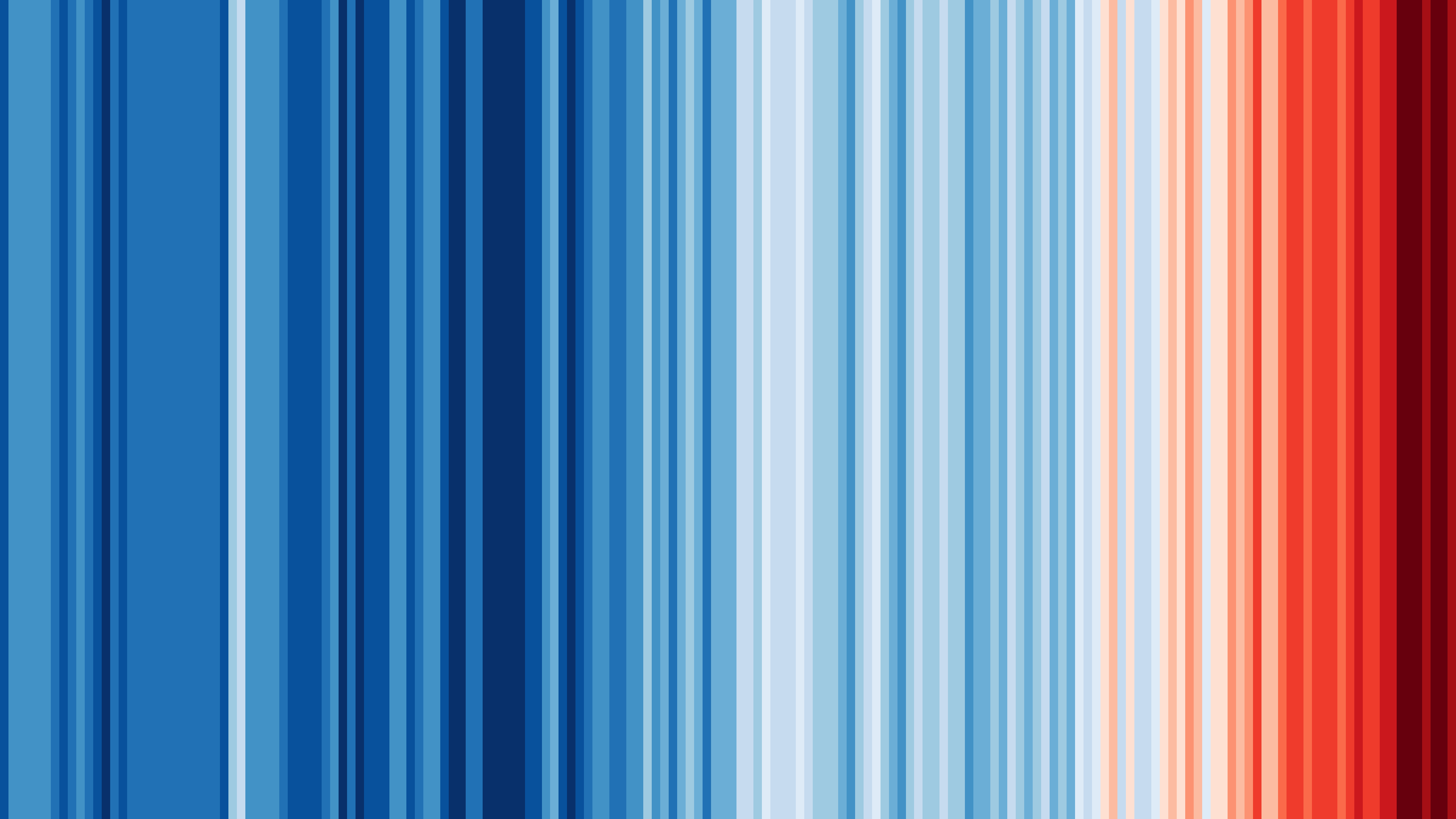 +
+
+
+**Graphics and Lead Scientist**: [Ed Hawkins](http://www.met.reading.ac.uk/~ed/home/index.php), National Centre for Atmospheric Science, University of Reading.
+
+**Data**: Berkeley Earth, NOAA, UK Met Office, MeteoSwiss, DWD, SMHI, UoR, Meteo France & ZAMG.
+
+
+#ShowYourStripes is distributed under a
+Creative Commons Attribution 4.0 International License
+
+  +
+
+
diff --git a/benchmarks/benchmarks/experimental/ugrid/__init__.py b/benchmarks/benchmarks/experimental/ugrid/__init__.py
index 2f9bb04e35..2e40c525a6 100644
--- a/benchmarks/benchmarks/experimental/ugrid/__init__.py
+++ b/benchmarks/benchmarks/experimental/ugrid/__init__.py
@@ -50,7 +50,7 @@ def time_create(self, *params):
class Connectivity(UGridCommon):
def setup(self, n_faces):
- self.array = np.zeros([n_faces, 3], dtype=np.int)
+ self.array = np.zeros([n_faces, 3], dtype=int)
super().setup(n_faces)
def create(self):
diff --git a/docs/Makefile b/docs/Makefile
index 47f3e740fa..fcb0ec0116 100644
--- a/docs/Makefile
+++ b/docs/Makefile
@@ -20,11 +20,6 @@ html-quick:
echo "make html-quick in $$i..."; \
(cd $$i; $(MAKE) $(MFLAGS) $(MYMAKEFLAGS) html-quick); done
-spelling:
- @for i in $(SUBDIRS); do \
- echo "make spelling in $$i..."; \
- (cd $$i; $(MAKE) $(MFLAGS) $(MYMAKEFLAGS) spelling); done
-
all:
@for i in $(SUBDIRS); do \
echo "make all in $$i..."; \
diff --git a/docs/gallery_code/general/plot_custom_aggregation.py b/docs/gallery_code/general/plot_custom_aggregation.py
index 5fba3669b6..6ef6075fb3 100644
--- a/docs/gallery_code/general/plot_custom_aggregation.py
+++ b/docs/gallery_code/general/plot_custom_aggregation.py
@@ -72,7 +72,7 @@ def main():
# Make an aggregator from the user function.
SPELL_COUNT = Aggregator(
- "spell_count", count_spells, units_func=lambda units: 1
+ "spell_count", count_spells, units_func=lambda units, **kwargs: 1
)
# Define the parameters of the test.
diff --git a/docs/gallery_code/general/plot_lineplot_with_legend.py b/docs/gallery_code/general/plot_lineplot_with_legend.py
index 78401817ba..aad7906acd 100644
--- a/docs/gallery_code/general/plot_lineplot_with_legend.py
+++ b/docs/gallery_code/general/plot_lineplot_with_legend.py
@@ -24,7 +24,6 @@ def main():
)
for cube in temperature.slices("longitude"):
-
# Create a string label to identify this cube (i.e. latitude: value).
cube_label = "latitude: %s" % cube.coord("latitude").points[0]
diff --git a/docs/gallery_code/general/plot_projections_and_annotations.py b/docs/gallery_code/general/plot_projections_and_annotations.py
index 75122591b9..2cf42e66e0 100644
--- a/docs/gallery_code/general/plot_projections_and_annotations.py
+++ b/docs/gallery_code/general/plot_projections_and_annotations.py
@@ -26,7 +26,6 @@
def make_plot(projection_name, projection_crs):
-
# Create a matplotlib Figure.
plt.figure()
diff --git a/docs/gallery_code/general/plot_zonal_means.py b/docs/gallery_code/general/plot_zonal_means.py
index 08a9578e63..195f8b4bb0 100644
--- a/docs/gallery_code/general/plot_zonal_means.py
+++ b/docs/gallery_code/general/plot_zonal_means.py
@@ -16,7 +16,6 @@
def main():
-
# Loads air_temp.pp and "collapses" longitude into a single, average value.
fname = iris.sample_data_path("air_temp.pp")
temperature = iris.load_cube(fname)
diff --git a/docs/gallery_code/meteorology/plot_lagged_ensemble.py b/docs/gallery_code/meteorology/plot_lagged_ensemble.py
index 5cd2752f39..e15aa0e6ef 100644
--- a/docs/gallery_code/meteorology/plot_lagged_ensemble.py
+++ b/docs/gallery_code/meteorology/plot_lagged_ensemble.py
@@ -86,7 +86,6 @@ def main():
# Iterate over all possible latitude longitude slices.
for cube in last_timestep.slices(["latitude", "longitude"]):
-
# Get the ensemble member number from the ensemble coordinate.
ens_member = cube.coord("realization").points[0]
diff --git a/docs/src/Makefile b/docs/src/Makefile
index 37c2e9e3e6..a75da5371b 100644
--- a/docs/src/Makefile
+++ b/docs/src/Makefile
@@ -62,11 +62,6 @@ html-quick:
@echo
@echo "Build finished. The HTML (no gallery or api docs) pages are in $(BUILDDIR)/html"
-spelling:
- $(SPHINXBUILD) -b spelling $(SRCDIR) $(BUILDDIR)
- @echo
- @echo "Build finished. The HTML (no gallery) pages are in $(BUILDDIR)/html"
-
dirhtml:
$(SPHINXBUILD) -b dirhtml $(ALLSPHINXOPTS) $(BUILDDIR)/dirhtml
@echo
@@ -156,4 +151,5 @@ doctest:
"results in $(BUILDDIR)/doctest/output.txt."
show:
- @python -c "import webbrowser; webbrowser.open_new_tab('file://$(shell pwd)/$(BUILDDIR)/html/index.html')"
\ No newline at end of file
+ @python -c "import webbrowser; webbrowser.open_new_tab('file://$(shell pwd)/$(BUILDDIR)/html/index.html')"
+
diff --git a/docs/src/common_links.inc b/docs/src/common_links.inc
index 530ebc4877..4d03a92715 100644
--- a/docs/src/common_links.inc
+++ b/docs/src/common_links.inc
@@ -9,7 +9,7 @@
.. _conda: https://docs.conda.io/en/latest/
.. _contributor: https://github.com/SciTools/scitools.org.uk/blob/master/contributors.json
.. _core developers: https://github.com/SciTools/scitools.org.uk/blob/master/contributors.json
-.. _generating sss keys for GitHub: https://docs.github.com/en/github/authenticating-to-github/adding-a-new-ssh-key-to-your-github-account
+.. _generating ssh keys for GitHub: https://docs.github.com/en/github/authenticating-to-github/adding-a-new-ssh-key-to-your-github-account
.. _GitHub Actions: https://docs.github.com/en/actions
.. _GitHub Help Documentation: https://docs.github.com/en/github
.. _GitHub Discussions: https://github.com/SciTools/iris/discussions
@@ -40,22 +40,26 @@
.. _CF-UGRID: https://ugrid-conventions.github.io/ugrid-conventions/
.. _issues on GitHub: https://github.com/SciTools/iris/issues?q=is%3Aopen+is%3Aissue+sort%3Areactions-%2B1-desc
.. _python-stratify: https://github.com/SciTools/python-stratify
+.. _iris-esmf-regrid: https://github.com/SciTools-incubator/iris-esmf-regrid
+.. _netCDF4: https://github.com/Unidata/netcdf4-python
.. comment
- Core developers (@github names) in alphabetical order:
+ Core developers and prolific contributors (@github names) in alphabetical order:
.. _@abooton: https://github.com/abooton
.. _@alastair-gemmell: https://github.com/alastair-gemmell
.. _@ajdawson: https://github.com/ajdawson
.. _@bjlittle: https://github.com/bjlittle
.. _@bouweandela: https://github.com/bouweandela
+.. _@bsherratt: https://github.com/bsherratt
.. _@corinnebosley: https://github.com/corinnebosley
.. _@cpelley: https://github.com/cpelley
.. _@djkirkham: https://github.com/djkirkham
.. _@DPeterK: https://github.com/DPeterK
.. _@ESadek-MO: https://github.com/ESadek-MO
.. _@esc24: https://github.com/esc24
+.. _@HGWright: https://github.com/HGWright
.. _@jamesp: https://github.com/jamesp
.. _@jonseddon: https://github.com/jonseddon
.. _@jvegasbsc: https://github.com/jvegasbsc
diff --git a/docs/src/community/index.rst b/docs/src/community/index.rst
new file mode 100644
index 0000000000..114cb96fe9
--- /dev/null
+++ b/docs/src/community/index.rst
@@ -0,0 +1,58 @@
+.. include:: ../common_links.inc
+
+.. todo:
+ consider scientific-python.org
+ consider scientific-python.org/specs/
+
+Iris in the Community
+=====================
+
+Iris aims to be a valuable member of the open source scientific Python
+community.
+
+We listen out for developments in our dependencies and neighbouring projects,
+and we reach out to them when we can solve problems together; please feel free
+to reach out to us!
+
+We are aware of our place in the user's wider 'toolbox' - offering unique
+functionality and interoperating smoothly with other packages.
+
+We welcome contributions from all; whether that's an opinion, a 1-line
+clarification, or a whole new feature 🙂
+
+Quick Links
+-----------
+
+* `GitHub Discussions`_
+* :ref:`Getting involved`
+* `Twitter `_
+
+Interoperability
+----------------
+
+There's a big choice of Python tools out there! Each one has strengths and
+weaknesses in different areas, so we don't want to force a single choice for your
+whole workflow - we'd much rather make it easy for you to choose the right tool
+for the moment, switching whenever you need. Below are our ongoing efforts at
+smoother interoperability:
+
+.. not using toctree due to combination of child pages and cross-references.
+
+* The :mod:`iris.pandas` module
+* :doc:`iris_xarray`
+
+.. toctree::
+ :maxdepth: 1
+ :hidden:
+
+ iris_xarray
+
+Plugins
+-------
+
+Iris can be extended with **plugins**! See below for further information:
+
+.. toctree::
+ :maxdepth: 2
+
+ plugins
diff --git a/docs/src/community/iris_xarray.rst b/docs/src/community/iris_xarray.rst
new file mode 100644
index 0000000000..859597da78
--- /dev/null
+++ b/docs/src/community/iris_xarray.rst
@@ -0,0 +1,154 @@
+.. include:: ../common_links.inc
+
+======================
+Iris ❤️ :term:`Xarray`
+======================
+
+There is a lot of overlap between Iris and :term:`Xarray`, but some important
+differences too. Below is a summary of the most important differences, so that
+you can be prepared, and to help you choose the best package for your use case.
+
+Overall Experience
+------------------
+
+Iris is the more specialised package, focussed on making it as easy
+as possible to work with meteorological and climatological data. Iris
+is built to natively handle many key concepts, such as the CF conventions,
+coordinate systems and bounded coordinates. Iris offers a smaller toolkit of
+operations compared to Xarray, particularly around API for sophisticated
+computation such as array manipulation and multi-processing.
+
+Xarray's more generic data model and community-driven development give it a
+richer range of operations and broader possible uses. Using Xarray
+specifically for meteorology/climatology may require deeper knowledge
+compared to using Iris, and you may prefer to add Xarray plugins
+such as :ref:`cfxarray` to get the best experience. Advanced users can likely
+achieve better performance with Xarray than with Iris.
+
+Conversion
+----------
+There are multiple ways to convert between Iris and Xarray objects.
+
+* Xarray includes the :meth:`~xarray.DataArray.to_iris` and
+ :meth:`~xarray.DataArray.from_iris` methods - detailed in the
+ `Xarray IO notes on Iris`_. Since Iris evolves independently of Xarray, be
+ vigilant for concepts that may be lost during the conversion.
+* Because both packages are closely linked to the :term:`NetCDF Format`, it is
+ feasible to save a NetCDF file using one package then load that file using
+ the other package. This will be lossy in places, as both Iris and Xarray
+ are opinionated on how certain NetCDF concepts relate to their data models.
+* The Iris development team are exploring an improved 'bridge' between the two
+ packages. Follow the conversation on GitHub: `iris#4994`_. This project is
+ expressly intended to be as lossless as possible.
+
+Regridding
+----------
+Iris and Xarray offer a range of regridding methods - both natively and via
+additional packages such as `iris-esmf-regrid`_ and `xESMF`_ - which overlap
+in places
+but tend to cover a different set of use cases (e.g. Iris handles unstructured
+meshes but offers access to fewer ESMF methods). The behaviour of these
+regridders also differs slightly (even between different regridders attached to
+the same package) so the appropriate package to use depends highly on the
+particulars of the use case.
+
+Plotting
+--------
+Xarray and Iris have a large overlap of functionality when creating
+:term:`Matplotlib` plots and both support the plotting of multidimensional
+coordinates. This means the experience is largely similar using either package.
+
+Xarray supports further plotting backends through external packages (e.g. Bokeh through `hvPlot`_)
+and, if a user is already familiar with `pandas`_, the interface should be
+familiar. It also supports some different plot types to Iris, and therefore can
+be used for a wider variety of plots. It also has benefits regarding "out of
+the box", quick customisations to plots. However, if further customisation is
+required, knowledge of matplotlib is still required.
+
+In both cases, :term:`Cartopy` is/can be used. Iris does more work
+automatically for the user here, creating Cartopy
+:class:`~cartopy.mpl.geoaxes.GeoAxes` for latitude and longitude coordinates,
+whereas the user has to do this manually in Xarray.
+
+Statistics
+----------
+Both libraries are quite comparable with generally similar capabilities,
+performance and laziness. Iris offers more specificity in some cases, such as
+some more specific unique functions and masked tolerance in most statistics.
+Xarray seems more approachable however, with some less unique but more
+convenient solutions (these tend to be wrappers to :term:`Dask` functions).
+
+Laziness and Multi-Processing with :term:`Dask`
+-----------------------------------------------
+Iris and Xarray both support lazy data and out-of-core processing through
+utilisation of Dask.
+
+While both Iris and Xarray expose :term:`NumPy` conveniences at the API level
+(e.g. the `ndim()` method), only Xarray exposes Dask conveniences. For example
+:attr:`xarray.DataArray.chunks`, which gives the user direct control
+over the underlying Dask array chunks. The Iris API instead takes control of
+such concepts and user control is only possible by manipulating the underlying
+Dask array directly (accessed via :meth:`iris.cube.Cube.core_data`).
+
+:class:`xarray.DataArray`\ s comply with `NEP-18`_, allowing NumPy arrays to be
+based on them, and they also include the necessary extra members for Dask
+arrays to be based on them too. Neither of these is currently possible with
+Iris :class:`~iris.cube.Cube`\ s, although an ambition for the future.
+
+NetCDF File Control
+-------------------
+(More info: :term:`NetCDF Format`)
+
+Unlike Iris, Xarray generally provides full control of major file structures,
+i.e. dimensions + variables, including their order in the file. It mostly
+respects these in a file input, and can reproduce them on output.
+However, attribute handling is not so complete: like Iris, it interprets and
+modifies some recognised aspects, and can add some extra attributes not in the
+input.
+
+.. todo:
+ More detail on dates and fill values (@pp-mo suggestion).
+
+Handling of dates and fill values have some special problems here.
+
+Ultimately, nearly everything wanted in a particular desired result file can
+be achieved in Xarray, via provided override mechanisms (`loading keywords`_
+and the '`encoding`_' dictionaries).
+
+Missing Data
+------------
+Xarray uses :data:`numpy.nan` to represent missing values and this will support
+many simple use cases assuming the data are floats. Iris enables more
+sophisticated missing data handling by representing missing values as masks
+(:class:`numpy.ma.MaskedArray` for real data and :class:`dask.array.Array`
+for lazy data) which allows data to be any data type and to include either/both
+a mask and :data:`~numpy.nan`\ s.
+
+.. _cfxarray:
+
+`cf-xarray`_
+-------------
+Iris has a data model entirely based on :term:`CF Conventions`. Xarray has a
+data model based on :term:`NetCDF Format` with cf-xarray acting as translation
+into CF. Xarray/cf-xarray methods can be
+called and data accessed with CF like arguments (e.g. axis, standard name) and
+there are some CF specific utilities (similar
+to Iris utilities). Iris tends to cover more of and be stricter about CF.
+
+
+.. seealso::
+
+ * `Xarray IO notes on Iris`_
+ * `Xarray notes on other NetCDF libraries`_
+
+.. _Xarray IO notes on Iris: https://docs.xarray.dev/en/stable/user-guide/io.html#iris
+.. _Xarray notes on other NetCDF libraries: https://docs.xarray.dev/en/stable/getting-started-guide/faq.html#what-other-netcdf-related-python-libraries-should-i-know-about
+.. _loading keywords: https://docs.xarray.dev/en/stable/generated/xarray.open_dataset.html#xarray.open_dataset
+.. _encoding: https://docs.xarray.dev/en/stable/user-guide/io.html#writing-encoded-data
+.. _xESMF: https://github.com/pangeo-data/xESMF/
+.. _seaborn: https://seaborn.pydata.org/
+.. _hvPlot: https://hvplot.holoviz.org/
+.. _pandas: https://pandas.pydata.org/
+.. _NEP-18: https://numpy.org/neps/nep-0018-array-function-protocol.html
+.. _cf-xarray: https://github.com/xarray-contrib/cf-xarray
+.. _iris#4994: https://github.com/SciTools/iris/issues/4994
diff --git a/docs/src/community/plugins.rst b/docs/src/community/plugins.rst
new file mode 100644
index 0000000000..0d79d64623
--- /dev/null
+++ b/docs/src/community/plugins.rst
@@ -0,0 +1,68 @@
+.. _namespace package: https://packaging.python.org/en/latest/guides/packaging-namespace-packages/
+
+.. _community_plugins:
+
+Plugins
+=======
+
+Iris supports **plugins** under the ``iris.plugins`` `namespace package`_.
+This allows packages that extend Iris' functionality to be developed and
+maintained independently, while still being installed into ``iris.plugins``
+instead of a separate package. For example, a plugin may provide loaders or
+savers for additional file formats, or alternative visualisation methods.
+
+
+Using plugins
+-------------
+
+Once a plugin is installed, it can be used either via the
+:func:`iris.use_plugin` function, or by importing it directly:
+
+.. code-block:: python
+
+ import iris
+
+ iris.use_plugin("my_plugin")
+ # OR
+ import iris.plugins.my_plugin
+
+
+Creating plugins
+----------------
+
+The choice of a `namespace package`_ makes writing a plugin relatively
+straightforward: it simply needs to appear as a folder within ``iris/plugins``,
+then can be distributed in the same way as any other package. An example
+repository layout:
+
+.. code-block:: text
+
+ + lib
+ + iris
+ + plugins
+ + my_plugin
+ - __init__.py
+ - (more code...)
+ - README.md
+ - pyproject.toml
+ - setup.cfg
+ - (other project files...)
+
+In particular, note that there must **not** be any ``__init__.py`` files at
+higher levels than the plugin itself.
+
+The package name - how it is referred to by PyPI/conda, specified by
+``metadata.name`` in ``setup.cfg`` - is recommended to include both "iris" and
+the plugin name. Continuing this example, its ``setup.cfg`` should include, at
+minimum:
+
+.. code-block:: ini
+
+ [metadata]
+ name = iris-my-plugin
+
+ [options]
+ packages = find_namespace:
+
+ [options.packages.find]
+ where = lib
diff --git a/docs/src/conf.py b/docs/src/conf.py
index 499d50116b..d2a204d3b3 100644
--- a/docs/src/conf.py
+++ b/docs/src/conf.py
@@ -158,9 +158,6 @@ def _dotv(version):
"sphinx_copybutton",
"sphinx.ext.napoleon",
"sphinx_panels",
- "sphinx_togglebutton",
- # TODO: Spelling extension disabled until the dependencies can be included
- # "sphinxcontrib.spelling",
"sphinx_gallery.gen_gallery",
"matplotlib.sphinxext.mathmpl",
"matplotlib.sphinxext.plot_directive",
@@ -199,16 +196,6 @@ def _dotv(version):
napoleon_use_keyword = True
napoleon_custom_sections = None
-# -- spellingextension --------------------------------------------------------
-# See https://sphinxcontrib-spelling.readthedocs.io/en/latest/customize.html
-spelling_lang = "en_GB"
-# The lines in this file must only use line feeds (no carriage returns).
-spelling_word_list_filename = ["spelling_allow.txt"]
-spelling_show_suggestions = False
-spelling_show_whole_line = False
-spelling_ignore_importable_modules = True
-spelling_ignore_python_builtins = True
-
# -- copybutton extension -----------------------------------------------------
# See https://sphinx-copybutton.readthedocs.io/en/latest/
copybutton_prompt_text = r">>> |\.\.\. "
@@ -241,6 +228,7 @@ def _dotv(version):
"python": ("https://docs.python.org/3/", None),
"scipy": ("https://docs.scipy.org/doc/scipy/", None),
"pandas": ("https://pandas.pydata.org/docs/", None),
+ "dask": ("https://docs.dask.org/en/stable/", None),
}
# The name of the Pygments (syntax highlighting) style to use.
@@ -289,7 +277,8 @@ def _dotv(version):
# See https://pydata-sphinx-theme.readthedocs.io/en/latest/user_guide/configuring.html
html_theme_options = {
- "footer_items": ["copyright", "sphinx-version", "custom_footer"],
+ "footer_start": ["copyright", "sphinx-version"],
+ "footer_end": ["custom_footer"],
"collapse_navigation": True,
"navigation_depth": 3,
"show_prev_next": True,
@@ -381,8 +370,6 @@ def _dotv(version):
"http://www.esrl.noaa.gov/psd/data/gridded/conventions/cdc_netcdf_standard.shtml",
"http://www.nationalarchives.gov.uk/doc/open-government-licence",
"https://www.metoffice.gov.uk/",
- # TODO: try removing this again in future - was raising an SSLError.
- "http://www.ecmwf.int/",
]
# list of sources to exclude from the build.
diff --git a/docs/src/developers_guide/contributing_documentation_full.rst b/docs/src/developers_guide/contributing_documentation_full.rst
index 390f2eeea7..a470def683 100755
--- a/docs/src/developers_guide/contributing_documentation_full.rst
+++ b/docs/src/developers_guide/contributing_documentation_full.rst
@@ -113,18 +113,6 @@ adding it to the ``linkcheck_ignore`` array that is defined in the
If this fails check the output for the text **broken** and then correct
or ignore the url.
-.. comment
- Finally, the spelling in the documentation can be checked automatically via the
- command::
-
- make spelling
-
- The spelling check may pull up many technical abbreviations and acronyms. This
- can be managed by using an **allow** list in the form of a file. This file,
- or list of files is set in the `conf.py`_ using the string list
- ``spelling_word_list_filename``.
-
-
.. note:: In addition to the automated `Iris GitHub Actions`_ build of all the
documentation build options above, the
https://readthedocs.org/ service is also used. The configuration
diff --git a/docs/src/developers_guide/gitwash/forking.rst b/docs/src/developers_guide/gitwash/forking.rst
index 247e3cf678..baeb243c86 100644
--- a/docs/src/developers_guide/gitwash/forking.rst
+++ b/docs/src/developers_guide/gitwash/forking.rst
@@ -18,7 +18,7 @@ Set up and Configure a Github Account
If you don't have a github account, go to the github page, and make one.
You then need to configure your account to allow write access, see
-the `generating sss keys for GitHub`_ help on `github help`_.
+the `generating ssh keys for GitHub`_ help on `github help`_.
Create Your own Forked Copy of Iris
diff --git a/docs/src/developers_guide/gitwash/set_up_fork.rst b/docs/src/developers_guide/gitwash/set_up_fork.rst
index d5c5bc5c44..5318825488 100644
--- a/docs/src/developers_guide/gitwash/set_up_fork.rst
+++ b/docs/src/developers_guide/gitwash/set_up_fork.rst
@@ -15,7 +15,7 @@ Overview
git clone git@github.com:your-user-name/iris.git
cd iris
- git remote add upstream git://github.com/SciTools/iris.git
+ git remote add upstream git@github.com/SciTools/iris.git
In Detail
=========
diff --git a/docs/src/developers_guide/release.rst b/docs/src/developers_guide/release.rst
index de7aa6c719..bae77a7d21 100644
--- a/docs/src/developers_guide/release.rst
+++ b/docs/src/developers_guide/release.rst
@@ -277,6 +277,11 @@ Post Release Steps
#. On main, make a new ``latest.rst`` from ``latest.rst.template`` and update
the include statement and the toctree in ``index.rst`` to point at the new
``latest.rst``.
+#. Consider updating ``docs/src/userguide/citation.rst`` on ``main`` to include
+ the version number, date and `Zenodo DOI `_
+ of the new release. Ideally this would be updated before the release, but
+ the DOI for the new version is only available once the release has been
+ created in GitHub.
.. _SciTools/iris: https://github.com/SciTools/iris
diff --git a/docs/src/further_topics/metadata.rst b/docs/src/further_topics/metadata.rst
index de1afb15af..4c55047d4c 100644
--- a/docs/src/further_topics/metadata.rst
+++ b/docs/src/further_topics/metadata.rst
@@ -389,10 +389,10 @@ instances. Normally, this would cause issues. For example,
.. doctest:: richer-metadata
- >>> simply = {"one": np.int(1), "two": np.array([1.0, 2.0])}
+ >>> simply = {"one": np.int32(1), "two": np.array([1.0, 2.0])}
>>> simply
{'one': 1, 'two': array([1., 2.])}
- >>> fruity = {"one": np.int(1), "two": np.array([1.0, 2.0])}
+ >>> fruity = {"one": np.int32(1), "two": np.array([1.0, 2.0])}
>>> fruity
{'one': 1, 'two': array([1., 2.])}
>>> simply == fruity
@@ -419,7 +419,7 @@ However, metadata class equality is rich enough to handle this eventuality,
>>> metadata1
CubeMetadata(standard_name='air_temperature', long_name=None, var_name='air_temperature', units=Unit('K'), attributes={'one': 1, 'two': array([1., 2.])}, cell_methods=(CellMethod(method='mean', coord_names=('time',), intervals=('6 hour',), comments=()),))
- >>> metadata2 = cube.metadata._replace(attributes={"one": np.int(1), "two": np.array([1000.0, 2000.0])})
+ >>> metadata2 = cube.metadata._replace(attributes={"one": np.int32(1), "two": np.array([1000.0, 2000.0])})
>>> metadata2
CubeMetadata(standard_name='air_temperature', long_name=None, var_name='air_temperature', units=Unit('K'), attributes={'one': 1, 'two': array([1000., 2000.])}, cell_methods=(CellMethod(method='mean', coord_names=('time',), intervals=('6 hour',), comments=()),))
>>> metadata1 == metadata2
diff --git a/docs/src/further_topics/ugrid/partner_packages.rst b/docs/src/further_topics/ugrid/partner_packages.rst
index 8e36f4ffc2..75b54b037f 100644
--- a/docs/src/further_topics/ugrid/partner_packages.rst
+++ b/docs/src/further_topics/ugrid/partner_packages.rst
@@ -1,3 +1,5 @@
+.. include:: ../../common_links.inc
+
.. _ugrid partners:
Iris' Mesh Partner Packages
@@ -97,4 +99,3 @@ Applications
.. _GeoVista: https://github.com/bjlittle/geovista
.. _PyVista: https://docs.pyvista.org/index.html
-.. _iris-esmf-regrid: https://github.com/SciTools-incubator/iris-esmf-regrid
diff --git a/docs/src/index.rst b/docs/src/index.rst
index c5d654ed31..531c0e0b26 100644
--- a/docs/src/index.rst
+++ b/docs/src/index.rst
@@ -136,6 +136,15 @@ The legacy support resources:
developers_guide/contributing_getting_involved
+.. toctree::
+ :caption: Community
+ :maxdepth: 1
+ :name: community_index
+ :hidden:
+
+ Community
+
+
.. toctree::
:caption: Iris API
:maxdepth: 1
diff --git a/docs/src/installing.rst b/docs/src/installing.rst
index 6a2d2f6131..b2481973c0 100644
--- a/docs/src/installing.rst
+++ b/docs/src/installing.rst
@@ -14,7 +14,7 @@ Subsystem for Linux). This is a great option to get started with Iris
for users and developers. Be aware that we do not currently test against
any WSL_ distributions.
-.. _WSL: https://docs.microsoft.com/en-us/windows/wsl/install-win10
+.. _WSL: https://learn.microsoft.com/en-us/windows/wsl/install
.. note:: Iris is currently supported and tested against |python_support|
running on Linux. We do not currently actively test on other
diff --git a/docs/src/spelling_allow.txt b/docs/src/spelling_allow.txt
deleted file mode 100644
index ed883ac3bf..0000000000
--- a/docs/src/spelling_allow.txt
+++ /dev/null
@@ -1,361 +0,0 @@
-Admin
-Albers
-Arakawa
-Arg
-Args
-Autoscale
-Biggus
-CF
-CI
-Cartopy
-Checklist
-Color
-Conda
-Constraining
-DAP
-Dask
-Debian
-Duchon
-EO
-Eos
-Exner
-Fieldsfile
-Fieldsfiles
-FillValue
-Gb
-GeogCS
-Hovmoller
-Jul
-Jun
-Jupyter
-Lanczos
-Mappables
-Matplotlib
-Mb
-Modeling
-Mollweide
-NetCDF
-Nino
-PPfield
-PPfields
-Perez
-Proj
-Quickplot
-Regrids
-Royer
-Scitools
-Scitools
-Sep
-Stehfest
-Steroegraphic
-Subsetting
-TestCodeFormat
-TestLicenseHeaders
-Torvalds
-Trans
-Trenberth
-Tri
-URIs
-URLs
-Ubuntu
-Ugrid
-Unidata
-Vol
-Vuuren
-Workflow
-Yury
-Zaytsev
-Zorder
-abf
-abl
-advection
-aggregator
-aggregators
-alphap
-ancils
-antimeridian
-ap
-arg
-args
-arithmetic
-arraylike
-atol
-auditable
-aux
-basemap
-behaviour
-betap
-bhulev
-biggus
-blev
-boolean
-boundpoints
-branchname
-broadcastable
-bugfix
-bugfixes
-builtin
-bulev
-carrée
-cartesian
-celsius
-center
-centrepoints
-cf
-cftime
-chunksizes
-ci
-clabel
-cmap
-cmpt
-codebase
-color
-colorbar
-colorbars
-complevel
-conda
-config
-constraining
-convertor
-coord
-coords
-cs
-datafiles
-datatype
-datetime
-datetimes
-ddof
-deepcopy
-deprecations
-der
-dewpoint
-dict
-dicts
-diff
-discontiguities
-discontiguous
-djf
-docstring
-docstrings
-doi
-dom
-dropdown
-dtype
-dtypes
-dx
-dy
-edgecolor
-endian
-endianness
-equirectangular
-eta
-etc
-fh
-fieldsfile
-fieldsfiles
-fileformat
-fileformats
-filename
-filenames
-filepath
-filespec
-fullname
-func
-geolocations
-github
-gregorian
-grib
-gribapi
-gridcell
-griddata
-gridlines
-hPa
-hashable
-hindcast
-hyperlink
-hyperlinks
-idiff
-ieee
-ifunc
-imagehash
-inc
-init
-inline
-inplace
-int
-interable
-interpolator
-ints
-io
-isosurfaces
-iterable
-jja
-jupyter
-kwarg
-kwargs
-landsea
-lat
-latlon
-latlons
-lats
-lbcode
-lbegin
-lbext
-lbfc
-lbft
-lblrec
-lbmon
-lbmond
-lbnrec
-lbrsvd
-lbtim
-lbuser
-lbvc
-lbyr
-lbyrd
-lh
-lhs
-linewidth
-linted
-linting
-lon
-lons
-lt
-mam
-markup
-matplotlib
-matplotlibrc
-max
-mdtol
-meaned
-mercator
-metadata
-min
-mpl
-nanmask
-nc
-ndarray
-neighbor
-ness
-netCDF
-netcdf
-netcdftime
-nimrod
-np
-nsigma
-numpy
-nx
-ny
-online
-orog
-paramId
-params
-parsable
-pcolormesh
-pdf
-placeholders
-plugin
-png
-proj
-ps
-pseudocolor
-pseudocolour
-pseudocoloured
-py
-pyplot
-quickplot
-rST
-rc
-rd
-reST
-reStructuredText
-rebase
-rebases
-rebasing
-regrid
-regridded
-regridder
-regridders
-regridding
-regrids
-rel
-repo
-repos
-reprojecting
-rh
-rhs
-rst
-rtol
-scipy
-scitools
-seekable
-setup
-sines
-sinh
-spec
-specs
-src
-ssh
-st
-stashcode
-stashcodes
-stats
-std
-stdout
-str
-subcube
-subcubes
-submodule
-submodules
-subsetting
-sys
-tanh
-tb
-testcases
-tgt
-th
-timepoint
-timestamp
-timesteps
-todo
-tol
-tos
-traceback
-travis
-tripolar
-tuple
-tuples
-txt
-udunits
-ufunc
-ugrid
-ukmo
-un
-unhandled
-unicode
-unittest
-unrotate
-unrotated
-uris
-url
-urls
-util
-var
-versioning
-vmax
-vmin
-waypoint
-waypoints
-whitespace
-wildcard
-wildcards
-windspeeds
-withnans
-workflow
-workflows
-xN
-xx
-xxx
-zeroth
-zlev
-zonal
diff --git a/docs/src/userguide/citation.rst b/docs/src/userguide/citation.rst
index 0a3a85fb89..1498b9dfe1 100644
--- a/docs/src/userguide/citation.rst
+++ b/docs/src/userguide/citation.rst
@@ -15,11 +15,12 @@ For example::
@manual{Iris,
author = {{Met Office}},
- title = {Iris: A Python package for analysing and visualising meteorological and oceanographic data sets},
- edition = {v1.2},
- year = {2010 - 2013},
+ title = {Iris: A powerful, format-agnostic, and community-driven Python package for analysing and visualising Earth science data },
+ edition = {v3.4},
+ year = {2010 - 2022},
address = {Exeter, Devon },
- url = {http://scitools.org.uk/}
+ url = {http://scitools.org.uk/},
+ doi = {10.5281/zenodo.7386117}
}
@@ -33,7 +34,7 @@ Suggested format::
For example::
- Iris. v1.2. 28-Feb-2013. Met Office. UK. https://github.com/SciTools/iris/archive/v1.2.0.tar.gz 01-03-2013
+ Iris. v3.4. 1-Dec-2022. Met Office. UK. https://doi.org/10.5281/zenodo.7386117 22-12-2022
********************
@@ -46,7 +47,7 @@ Suggested format::
For example::
- Iris. Met Office. git@github.com:SciTools/iris.git 06-03-2013
+ Iris. Met Office. git@github.com:SciTools/iris.git 22-12-2022
.. _How to cite and describe software: https://software.ac.uk/how-cite-software
diff --git a/docs/src/userguide/cube_maths.rst b/docs/src/userguide/cube_maths.rst
index 9c0898b62c..56a2041bd3 100644
--- a/docs/src/userguide/cube_maths.rst
+++ b/docs/src/userguide/cube_maths.rst
@@ -5,8 +5,8 @@ Cube Maths
==========
-The section :doc:`navigating_a_cube` highlighted that
-every cube has a data attribute;
+The section :doc:`navigating_a_cube` highlighted that
+every cube has a data attribute;
this attribute can then be manipulated directly::
cube.data -= 273.15
@@ -37,7 +37,7 @@ Let's load some air temperature which runs from 1860 to 2100::
filename = iris.sample_data_path('E1_north_america.nc')
air_temp = iris.load_cube(filename, 'air_temperature')
-We can now get the first and last time slices using indexing
+We can now get the first and last time slices using indexing
(see :ref:`cube_indexing` for a reminder)::
t_first = air_temp[0, :, :]
@@ -50,8 +50,8 @@ We can now get the first and last time slices using indexing
t_first = air_temp[0, :, :]
t_last = air_temp[-1, :, :]
-And finally we can subtract the two.
-The result is a cube of the same size as the original two time slices,
+And finally we can subtract the two.
+The result is a cube of the same size as the original two time slices,
but with the data representing their difference:
>>> print(t_last - t_first)
@@ -70,8 +70,8 @@ but with the data representing their difference:
.. note::
- Notice that the coordinates "time" and "forecast_period" have been removed
- from the resultant cube;
+ Notice that the coordinates "time" and "forecast_period" have been removed
+ from the resultant cube;
this is because these coordinates differed between the two input cubes.
@@ -174,15 +174,15 @@ broadcasting behaviour::
Combining Multiple Phenomena to Form a New One
----------------------------------------------
-Combining cubes of potential-temperature and pressure we can calculate
+Combining cubes of potential-temperature and pressure we can calculate
the associated temperature using the equation:
.. math::
-
+
T = \theta (\frac{p}{p_0}) ^ {(287.05 / 1005)}
-Where :math:`p` is pressure, :math:`\theta` is potential temperature,
-:math:`p_0` is the potential temperature reference pressure
+Where :math:`p` is pressure, :math:`\theta` is potential temperature,
+:math:`p_0` is the potential temperature reference pressure
and :math:`T` is temperature.
First, let's load pressure and potential temperature cubes::
@@ -191,7 +191,7 @@ First, let's load pressure and potential temperature cubes::
phenomenon_names = ['air_potential_temperature', 'air_pressure']
pot_temperature, pressure = iris.load_cubes(filename, phenomenon_names)
-In order to calculate :math:`\frac{p}{p_0}` we can define a coordinate which
+In order to calculate :math:`\frac{p}{p_0}` we can define a coordinate which
represents the standard reference pressure of 1000 hPa::
import iris.coords
@@ -205,7 +205,7 @@ the :meth:`iris.coords.Coord.convert_units` method::
p0.convert_units(pressure.units)
-Now we can combine all of this information to calculate the air temperature
+Now we can combine all of this information to calculate the air temperature
using the equation above::
temperature = pot_temperature * ( (pressure / p0) ** (287.05 / 1005) )
@@ -219,12 +219,12 @@ The result could now be plotted using the guidance provided in the
.. only:: html
- A very similar example to this can be found in
+ A very similar example to this can be found in
:ref:`sphx_glr_generated_gallery_meteorology_plot_deriving_phenomena.py`.
.. only:: latex
- A very similar example to this can be found in the examples section,
+ A very similar example to this can be found in the examples section,
with the title "Deriving Exner Pressure and Air Temperature".
.. _cube_maths_combining_units:
@@ -249,7 +249,7 @@ unit (if ``a`` had units ``'m2'`` then ``a ** 0.5`` would result in a cube
with units ``'m'``).
Iris inherits units from `cf_units `_
-which in turn inherits from `UDUNITS `_.
+which in turn inherits from `UDUNITS `_.
As well as the units UDUNITS provides, cf units also provides the units
``'no-unit'`` and ``'unknown'``. A unit of ``'no-unit'`` means that the
associated data is not suitable for describing with a unit, cf units
diff --git a/docs/src/userguide/cube_statistics.rst b/docs/src/userguide/cube_statistics.rst
index 08297c2a51..9dc21f91b5 100644
--- a/docs/src/userguide/cube_statistics.rst
+++ b/docs/src/userguide/cube_statistics.rst
@@ -14,7 +14,7 @@ Cube Statistics
Collapsing Entire Data Dimensions
---------------------------------
-.. testsetup::
+.. testsetup:: collapsing
import iris
filename = iris.sample_data_path('uk_hires.pp')
@@ -125,7 +125,7 @@ in order to calculate the area of the grid boxes::
These areas can now be passed to the ``collapsed`` method as weights:
-.. doctest::
+.. doctest:: collapsing
>>> new_cube = cube.collapsed(['grid_longitude', 'grid_latitude'], iris.analysis.MEAN, weights=grid_areas)
>>> print(new_cube)
@@ -141,8 +141,8 @@ These areas can now be passed to the ``collapsed`` method as weights:
altitude - x
Scalar coordinates:
forecast_reference_time 2009-11-19 04:00:00
- grid_latitude 1.5145501 degrees, bound=(0.14430022, 2.8848) degrees
- grid_longitude 358.74948 degrees, bound=(357.494, 360.00497) degrees
+ grid_latitude 1.5145501 degrees, bound=(0.13755022, 2.89155) degrees
+ grid_longitude 358.74948 degrees, bound=(357.48724, 360.01172) degrees
surface_altitude 399.625 m, bound=(-14.0, 813.25) m
Cell methods:
mean grid_longitude, grid_latitude
@@ -155,6 +155,50 @@ Several examples of area averaging exist in the gallery which may be of interest
including an example on taking a :ref:`global area-weighted mean
`.
+In addition to plain arrays, weights can also be given as cubes or (names of)
+:meth:`~iris.cube.Cube.coords`, :meth:`~iris.cube.Cube.cell_measures`, or
+:meth:`~iris.cube.Cube.ancillary_variables`.
+This has the advantage of correct unit handling, e.g., for area-weighted sums
+the units of the resulting cube are multiplied by an area unit:
+
+.. doctest:: collapsing
+
+ >>> from iris.coords import CellMeasure
+ >>> cell_areas = CellMeasure(
+ ... grid_areas,
+ ... standard_name='cell_area',
+ ... units='m2',
+ ... measure='area',
+ ... )
+ >>> cube.add_cell_measure(cell_areas, (0, 1, 2, 3))
+ >>> area_weighted_sum = cube.collapsed(
+ ... ['grid_longitude', 'grid_latitude'],
+ ... iris.analysis.SUM,
+ ... weights='cell_area'
+ ... )
+ >>> print(area_weighted_sum)
+ air_potential_temperature / (m2.K) (time: 3; model_level_number: 7)
+ Dimension coordinates:
+ time x -
+ model_level_number - x
+ Auxiliary coordinates:
+ forecast_period x -
+ level_height - x
+ sigma - x
+ Derived coordinates:
+ altitude - x
+ Scalar coordinates:
+ forecast_reference_time 2009-11-19 04:00:00
+ grid_latitude 1.5145501 degrees, bound=(0.13755022, 2.89155) degrees
+ grid_longitude 358.74948 degrees, bound=(357.48724, 360.01172) degrees
+ surface_altitude 399.625 m, bound=(-14.0, 813.25) m
+ Cell methods:
+ sum grid_longitude, grid_latitude
+ Attributes:
+ STASH m01s00i004
+ source 'Data from Met Office Unified Model'
+ um_version '7.3'
+
.. _cube-statistics-aggregated-by:
Partially Reducing Data Dimensions
@@ -338,3 +382,44 @@ from jja-2006 to jja-2010:
mam 2010
jja 2010
+Moreover, :meth:`Cube.aggregated_by ` supports
+weighted aggregation.
+For example, this is helpful for an aggregation over a monthly time
+coordinate that consists of months with different numbers of days.
+Similar to :meth:`Cube.collapsed `, weights can be
+given as arrays, cubes, or as (names of) :meth:`~iris.cube.Cube.coords`,
+:meth:`~iris.cube.Cube.cell_measures`, or
+:meth:`~iris.cube.Cube.ancillary_variables`.
+When weights are not given as arrays, units are correctly handled for weighted
+sums, i.e., the original unit of the cube is multiplied by the units of the
+weights.
+The following example shows a weighted sum (notice the change of the units):
+
+.. doctest:: aggregation
+
+ >>> from iris.coords import AncillaryVariable
+ >>> time_weights = AncillaryVariable(
+ ... cube.coord("time").bounds[:, 1] - cube.coord("time").bounds[:, 0],
+ ... long_name="Time Weights",
+ ... units="hours",
+ ... )
+ >>> cube.add_ancillary_variable(time_weights, 0)
+ >>> seasonal_sum = cube.aggregated_by("clim_season", iris.analysis.SUM, weights="Time Weights")
+ >>> print(seasonal_sum)
+ surface_temperature / (3600 s.K) (-- : 4; latitude: 18; longitude: 432)
+ Dimension coordinates:
+ latitude - x -
+ longitude - - x
+ Auxiliary coordinates:
+ clim_season x - -
+ forecast_reference_time x - -
+ season_year x - -
+ time x - -
+ Scalar coordinates:
+ forecast_period 0 hours
+ Cell methods:
+ mean month, year
+ sum clim_season
+ Attributes:
+ Conventions 'CF-1.5'
+ STASH m01s00i024
diff --git a/docs/src/userguide/glossary.rst b/docs/src/userguide/glossary.rst
index 818ef0c7ad..5c24f03372 100644
--- a/docs/src/userguide/glossary.rst
+++ b/docs/src/userguide/glossary.rst
@@ -1,3 +1,5 @@
+.. include:: ../common_links.inc
+
.. _glossary:
Glossary
@@ -125,7 +127,7 @@ Glossary
of formats.
| **Related:** :term:`CartoPy` **|** :term:`NumPy`
- | **More information:** `Matplotlib `_
+ | **More information:** `matplotlib`_
|
Metadata
@@ -143,9 +145,11 @@ Glossary
When Iris loads this format, it also especially recognises and interprets data
encoded according to the :term:`CF Conventions`.
+ __ `NetCDF4`_
+
| **Related:** :term:`Fields File (FF) Format`
**|** :term:`GRIB Format` **|** :term:`Post Processing (PP) Format`
- | **More information:** `NetCDF-4 Python Git `_
+ | **More information:** `NetCDF-4 Python Git`__
|
NumPy
diff --git a/docs/src/userguide/plotting_examples/1d_with_legend.py b/docs/src/userguide/plotting_examples/1d_with_legend.py
index 9b9fd8a49d..626335af45 100644
--- a/docs/src/userguide/plotting_examples/1d_with_legend.py
+++ b/docs/src/userguide/plotting_examples/1d_with_legend.py
@@ -13,7 +13,6 @@
temperature = temperature[5:9, :]
for cube in temperature.slices("longitude"):
-
# Create a string label to identify this cube (i.e. latitude: value)
cube_label = "latitude: %s" % cube.coord("latitude").points[0]
diff --git a/docs/src/whatsnew/1.0.rst b/docs/src/whatsnew/1.0.rst
index b226dc609b..c256c33566 100644
--- a/docs/src/whatsnew/1.0.rst
+++ b/docs/src/whatsnew/1.0.rst
@@ -147,8 +147,7 @@ the surface pressure. In return, it provides a virtual "pressure"
coordinate whose values are derived from the given components.
This facility is utilised by the GRIB2 loader to automatically provide
-the derived "pressure" coordinate for certain data [#f1]_ from the
-`ECMWF `_.
+the derived "pressure" coordinate for certain data [#f1]_ from the ECMWF.
.. [#f1] Where the level type is either 105 or 119, and where the
surface pressure has an ECMWF paramId of
diff --git a/docs/src/whatsnew/2.1.rst b/docs/src/whatsnew/2.1.rst
index 18c562d3da..33f3a013b1 100644
--- a/docs/src/whatsnew/2.1.rst
+++ b/docs/src/whatsnew/2.1.rst
@@ -1,3 +1,5 @@
+.. include:: ../common_links.inc
+
v2.1 (06 Jun 2018)
******************
@@ -67,7 +69,7 @@ Incompatible Changes
as an alternative.
* This release of Iris contains a number of updated metadata translations.
- See this
+ See this
`changelist `_
for further information.
@@ -84,7 +86,7 @@ Internal
calendar.
* Iris updated its time-handling functionality from the
- `netcdf4-python `_
+ `netcdf4-python`__
``netcdftime`` implementation to the standalone module
`cftime `_.
cftime is entirely compatible with netcdftime, but some issues may
@@ -92,6 +94,8 @@ Internal
In this situation, simply replacing ``netcdftime.datetime`` with
``cftime.datetime`` should be sufficient.
+__ `netCDF4`_
+
* Iris now requires version 2 of Matplotlib, and ``>=1.14`` of NumPy.
Full requirements can be seen in the `requirements `_
directory of the Iris' the source.
diff --git a/docs/src/whatsnew/3.4.rst b/docs/src/whatsnew/3.4.rst
index 1ad676c049..02fc574e51 100644
--- a/docs/src/whatsnew/3.4.rst
+++ b/docs/src/whatsnew/3.4.rst
@@ -26,15 +26,29 @@ This document explains the changes made to Iris for this release
* We have **begun refactoring Iris' regridding**, which has already improved
performance and functionality, with more potential in future!
* We have made several other significant `🚀 Performance Enhancements`_.
- * Please note that **Iris cannot currently work with the latest NetCDF4
- releases**. The pin is set to ``` if you have
any issues or feature requests for improving Iris. Enjoy!
+v3.4.1 (21 Feb 2023)
+====================
+
+.. dropdown:: :opticon:`alert` v3.4.1 Patches
+ :container: + shadow
+ :title: text-primary text-center font-weight-bold
+ :body: bg-light
+ :animate: fade-in
+
+ The patches in this release of Iris include:
+
+ #. `@trexfeathers`_ and `@pp-mo`_ made Iris' use of the `netCDF4`_ library
+ thread-safe. (:pull:`5095`)
+
+ #. `@trexfeathers`_ and `@pp-mo`_ removed the netCDF4 pin mentioned in
+ `🔗 Dependencies`_ point 3. (:pull:`5095`)
+
+
📢 Announcements
================
diff --git a/docs/src/whatsnew/latest.rst b/docs/src/whatsnew/latest.rst
index 22ec7a00b8..92153f902c 100644
--- a/docs/src/whatsnew/latest.rst
+++ b/docs/src/whatsnew/latest.rst
@@ -10,7 +10,7 @@ This document explains the changes made to Iris for this release
The highlights for this major/minor release of Iris include:
- * N/A
+ * We're so proud to fully support `@ed-hawkins`_ and `#ShowYourStripes`_ ❤️
And finally, get in touch with us on :issue:`GitHub` if you have
any issues or feature requests for improving Iris. Enjoy!
@@ -20,18 +20,35 @@ This document explains the changes made to Iris for this release
================
#. Congratulations to `@ESadek-MO`_ who has become a core developer for Iris! 🎉
+#. Welcome and congratulations to `@HGWright`_ for making his first contribution to Iris! 🎉
✨ Features
===========
-#. N/A
+#. `@bsherratt`_ added support for plugins - see the corresponding
+ :ref:`documentation page` for further information.
+ (:pull:`5144`)
+
+#. `@rcomer`_ enabled lazy evaluation of :obj:`~iris.analysis.RMS` calcuations
+ with weights. (:pull:`5017`)
+
+#. `@schlunma`_ allowed the usage of cubes, coordinates, cell measures, or
+ ancillary variables as weights for cube aggregations
+ (:meth:`iris.cube.Cube.collapsed`, :meth:`iris.cube.Cube.aggregated_by`, and
+ :meth:`iris.cube.Cube.rolling_window`). This automatically adapts cube units
+ if necessary. (:pull:`5084`)
🐛 Bugs Fixed
=============
-#. N/A
+#. `@trexfeathers`_ and `@pp-mo`_ made Iris' use of the `netCDF4`_ library
+ thread-safe. (:pull:`5095`)
+
+#. `@ESadek-MO`_ removed check and error raise for saving
+ cubes with masked :class:`iris.coords.CellMeasure`.
+ (:issue:`5147`, :pull:`5181`)
💣 Incompatible Changes
@@ -66,10 +83,29 @@ This document explains the changes made to Iris for this release
the light version (not dark) while we make the docs dark mode friendly
(:pull:`5129`)
+#. `@jonseddon`_ updated the citation to a more recent version of Iris. (:pull:`5116`)
+
#. `@rcomer`_ linked the :obj:`~iris.analysis.PERCENTILE` aggregator from the
:obj:`~iris.analysis.MEDIAN` docstring, noting that the former handles lazy
data. (:pull:`5128`)
+#. `@trexfeathers`_ updated the WSL link to Microsoft's latest documentation,
+ and removed an ECMWF link in the ``v1.0`` What's New that was failing the
+ linkcheck CI. (:pull:`5109`)
+
+#. `@trexfeathers`_ added a new top-level :doc:`/community/index` section,
+ as a one-stop place to find out about getting involved, and how we relate
+ to other projects. (:pull:`5025`)
+
+#. The **Iris community**, with help from the **Xarray community**, produced
+ the :doc:`/community/iris_xarray` page, highlighting the similarities and
+ differences between the two packages. (:pull:`5025`)
+
+#. `@bjlittle`_ added a new section to the `README.md`_ to show our support
+ for the outstanding work of `@ed-hawkins`_ et al for `#ShowYourStripes`_.
+ (:pull:`5141`)
+
+#. `@HGWright`_ fixed some typo's from Gitwash. (:pull:`5145`)
💼 Internal
===========
@@ -80,14 +116,22 @@ This document explains the changes made to Iris for this release
#. `@rcomer`_ removed some old infrastructure that printed test timings.
(:pull:`5101`)
+#. `@lbdreyer`_ and `@trexfeathers`_ (reviewer) added coverage testing. This
+ can be enabled by using the "--coverage" flag when running the tests with
+ nox i.e. ``nox --session tests -- --coverage``. (:pull:`4765`)
+
+#. `@lbdreyer`_ and `@trexfeathers`_ (reviewer) removed the ``--coding-tests``
+ option from Iris' test runner. (:pull:`4765`)
.. comment
Whatsnew author names (@github name) in alphabetical order. Note that,
core dev names are automatically included by the common_links.inc:
.. _@fnattino: https://github.com/fnattino
-
+.. _@ed-hawkins: https://github.com/ed-hawkins
.. comment
Whatsnew resources in alphabetical order:
+.. _#ShowYourStripes: https://showyourstripes.info/s/globe/
+.. _README.md: https://github.com/SciTools/iris#-----
diff --git a/lib/iris/__init__.py b/lib/iris/__init__.py
index 896b850541..38465472ee 100644
--- a/lib/iris/__init__.py
+++ b/lib/iris/__init__.py
@@ -91,6 +91,7 @@ def callback(cube, field, filename):
import contextlib
import glob
+import importlib
import itertools
import os.path
import pathlib
@@ -129,6 +130,7 @@ def callback(cube, field, filename):
"sample_data_path",
"save",
"site_configuration",
+ "use_plugin",
]
@@ -175,7 +177,6 @@ def __init__(self, datum_support=False, pandas_ndim=False):
self.__dict__["pandas_ndim"] = pandas_ndim
def __repr__(self):
-
# msg = ('Future(example_future_flag={})')
# return msg.format(self.example_future_flag)
msg = "Future(datum_support={}, pandas_ndim={})"
@@ -471,3 +472,22 @@ def sample_data_path(*path_to_join):
"appropriate for general file access.".format(target)
)
return target
+
+
+def use_plugin(plugin_name):
+ """
+ Convenience function to import a plugin
+
+ For example::
+
+ use_plugin("my_plugin")
+
+ is equivalent to::
+
+ import iris.plugins.my_plugin
+
+ This is useful for plugins that are not used directly, but instead do all
+ their setup on import. In this case, style checkers would not know the
+ significance of the import statement and warn that it is an unused import.
+ """
+ importlib.import_module(f"iris.plugins.{plugin_name}")
diff --git a/lib/iris/_lazy_data.py b/lib/iris/_lazy_data.py
index ac7ae34511..e0566fc8f2 100644
--- a/lib/iris/_lazy_data.py
+++ b/lib/iris/_lazy_data.py
@@ -39,7 +39,7 @@ def is_lazy_data(data):
"""
Return whether the argument is an Iris 'lazy' data array.
- At present, this means simply a Dask array.
+ At present, this means simply a :class:`dask.array.Array`.
We determine this by checking for a "compute" property.
"""
@@ -67,7 +67,8 @@ def _optimum_chunksize_internals(
* shape (tuple of int):
The full array shape of the target data.
* limit (int):
- The 'ideal' target chunk size, in bytes. Default from dask.config.
+ The 'ideal' target chunk size, in bytes. Default from
+ :mod:`dask.config`.
* dtype (np.dtype):
Numpy dtype of target data.
@@ -77,7 +78,7 @@ def _optimum_chunksize_internals(
.. note::
The purpose of this is very similar to
- `dask.array.core.normalize_chunks`, when called as
+ :func:`dask.array.core.normalize_chunks`, when called as
`(chunks='auto', shape, dtype=dtype, previous_chunks=chunks, ...)`.
Except, the operation here is optimised specifically for a 'c-like'
dimension order, i.e. outer dimensions first, as for netcdf variables.
@@ -174,13 +175,13 @@ def _optimum_chunksize(
def as_lazy_data(data, chunks=None, asarray=False):
"""
- Convert the input array `data` to a dask array.
+ Convert the input array `data` to a :class:`dask.array.Array`.
Args:
* data (array-like):
An indexable object with 'shape', 'dtype' and 'ndim' properties.
- This will be converted to a dask array.
+ This will be converted to a :class:`dask.array.Array`.
Kwargs:
@@ -192,7 +193,7 @@ def as_lazy_data(data, chunks=None, asarray=False):
Set to False (default) to pass passed chunks through unchanged.
Returns:
- The input array converted to a dask array.
+ The input array converted to a :class:`dask.array.Array`.
.. note::
The result chunk size is a multiple of 'chunks', if given, up to the
@@ -284,15 +285,16 @@ def multidim_lazy_stack(stack):
"""
Recursively build a multidimensional stacked dask array.
- This is needed because dask.array.stack only accepts a 1-dimensional list.
+ This is needed because :meth:`dask.array.Array.stack` only accepts a
+ 1-dimensional list.
Args:
* stack:
- An ndarray of dask arrays.
+ An ndarray of :class:`dask.array.Array`.
Returns:
- The input array converted to a lazy dask array.
+ The input array converted to a lazy :class:`dask.array.Array`.
"""
if stack.ndim == 0:
diff --git a/lib/iris/_merge.py b/lib/iris/_merge.py
index bc12080523..5ca5f31a8e 100644
--- a/lib/iris/_merge.py
+++ b/lib/iris/_merge.py
@@ -1418,6 +1418,7 @@ def _define_space(self, space, positions, indexes, function_matrix):
participates in a functional relationship.
"""
+
# Heuristic reordering of coordinate defintion indexes into
# preferred dimension order.
def axis_and_name(name):
diff --git a/lib/iris/analysis/__init__.py b/lib/iris/analysis/__init__.py
index 55d5d5d93e..173487cfb0 100644
--- a/lib/iris/analysis/__init__.py
+++ b/lib/iris/analysis/__init__.py
@@ -39,8 +39,10 @@
from collections.abc import Iterable
import functools
from functools import wraps
+from inspect import getfullargspec
import warnings
+from cf_units import Unit
import dask.array as da
import numpy as np
import numpy.ma as ma
@@ -55,7 +57,9 @@
)
from iris.analysis._regrid import CurvilinearRegridder, RectilinearRegridder
import iris.coords
+from iris.coords import _DimensionalMetadata
from iris.exceptions import LazyAggregatorError
+import iris.util
__all__ = (
"Aggregator",
@@ -296,7 +300,6 @@ def _dimensional_metadata_comparison(*cubes, object_get=None):
# for coordinate groups
for cube, coords in zip(cubes, all_coords):
for coord in coords:
-
# if this coordinate has already been processed, then continue on
# to the next one
if id(coord) in processed_coords:
@@ -468,11 +471,13 @@ def __init__(
Kwargs:
* units_func (callable):
- | *Call signature*: (units)
+ | *Call signature*: (units, \**kwargs)
If provided, called to convert a cube's units.
Returns an :class:`cf_units.Unit`, or a
value that can be made into one.
+ To ensure backwards-compatibility, also accepts a callable with
+ call signature (units).
* lazy_func (callable or None):
An alternative to :data:`call_func` implementing a lazy
@@ -480,7 +485,8 @@ def __init__(
main operation, but should raise an error in unhandled cases.
Additional kwargs::
- Passed through to :data:`call_func` and :data:`lazy_func`.
+ Passed through to :data:`call_func`, :data:`lazy_func`, and
+ :data:`units_func`.
Aggregators are used by cube aggregation methods such as
:meth:`~iris.cube.Cube.collapsed` and
@@ -626,7 +632,11 @@ def update_metadata(self, cube, coords, **kwargs):
"""
# Update the units if required.
if self.units_func is not None:
- cube.units = self.units_func(cube.units)
+ argspec = getfullargspec(self.units_func)
+ if argspec.varkw is None: # old style
+ cube.units = self.units_func(cube.units)
+ else: # new style (preferred)
+ cube.units = self.units_func(cube.units, **kwargs)
def post_process(self, collapsed_cube, data_result, coords, **kwargs):
"""
@@ -694,13 +704,13 @@ class PercentileAggregator(_Aggregator):
"""
def __init__(self, units_func=None, **kwargs):
- """
+ r"""
Create a percentile aggregator.
Kwargs:
* units_func (callable):
- | *Call signature*: (units)
+ | *Call signature*: (units, \**kwargs)
If provided, called to convert a cube's units.
Returns an :class:`cf_units.Unit`, or a
@@ -935,13 +945,13 @@ class WeightedPercentileAggregator(PercentileAggregator):
"""
def __init__(self, units_func=None, lazy_func=None, **kwargs):
- """
+ r"""
Create a weighted percentile aggregator.
Kwargs:
* units_func (callable):
- | *Call signature*: (units)
+ | *Call signature*: (units, \**kwargs)
If provided, called to convert a cube's units.
Returns an :class:`cf_units.Unit`, or a
@@ -1173,8 +1183,112 @@ def post_process(self, collapsed_cube, data_result, coords, **kwargs):
return result
+class _Weights(np.ndarray):
+ """Class for handling weights for weighted aggregation.
+
+ This subclasses :class:`numpy.ndarray`; thus, all methods and properties of
+ :class:`numpy.ndarray` (e.g., `shape`, `ndim`, `view()`, etc.) are
+ available.
+
+ Details on subclassing :class:`numpy.ndarray` are given here:
+ https://numpy.org/doc/stable/user/basics.subclassing.html
+
+ """
+
+ def __new__(cls, weights, cube, units=None):
+ """Create class instance.
+
+ Args:
+
+ * weights (Cube, string, _DimensionalMetadata, array-like):
+ If given as a :class:`iris.cube.Cube`, use its data and units. If
+ given as a :obj:`str` or :class:`iris.coords._DimensionalMetadata`,
+ assume this is (the name of) a
+ :class:`iris.coords._DimensionalMetadata` object of the cube (i.e.,
+ one of :meth:`iris.cube.Cube.coords`,
+ :meth:`iris.cube.Cube.cell_measures`, or
+ :meth:`iris.cube.Cube.ancillary_variables`). If given as an
+ array-like object, use this directly and assume units of `1`. If
+ `units` is given, ignore all units derived above and use the ones
+ given by `units`.
+ * cube (Cube):
+ Input cube for aggregation. If weights is given as :obj:`str` or
+ :class:`iris.coords._DimensionalMetadata`, try to extract the
+ :class:`iris.coords._DimensionalMetadata` object and corresponding
+ dimensional mappings from this cube. Otherwise, this argument is
+ ignored.
+ * units (string, Unit):
+ If ``None``, use units derived from `weights`. Otherwise, overwrite
+ the units derived from `weights` and use `units`.
+
+ """
+ # `weights` is a cube
+ # Note: to avoid circular imports of Cube we use duck typing using the
+ # "hasattr" syntax here
+ # --> Extract data and units from cube
+ if hasattr(weights, "add_aux_coord"):
+ obj = np.asarray(weights.data).view(cls)
+ obj.units = weights.units
+
+ # `weights`` is a string or _DimensionalMetadata object
+ # --> Extract _DimensionalMetadata object from cube, broadcast it to
+ # correct shape using the corresponding dimensional mapping, and use
+ # its data and units
+ elif isinstance(weights, (str, _DimensionalMetadata)):
+ dim_metadata = cube._dimensional_metadata(weights)
+ arr = dim_metadata._values
+ if dim_metadata.shape != cube.shape:
+ arr = iris.util.broadcast_to_shape(
+ arr,
+ cube.shape,
+ dim_metadata.cube_dims(cube),
+ )
+ obj = np.asarray(arr).view(cls)
+ obj.units = dim_metadata.units
+
+ # Remaining types (e.g., np.ndarray): try to convert to ndarray.
+ else:
+ obj = np.asarray(weights).view(cls)
+ obj.units = Unit("1")
+
+ # Overwrite units from units argument if necessary
+ if units is not None:
+ obj.units = units
+
+ return obj
+
+ def __array_finalize__(self, obj):
+ """See https://numpy.org/doc/stable/user/basics.subclassing.html.
+
+ Note
+ ----
+ `obj` cannot be `None` here since ``_Weights.__new__`` does not call
+ ``super().__new__`` explicitly.
+
+ """
+ self.units = getattr(obj, "units", Unit("1"))
+
+ @classmethod
+ def update_kwargs(cls, kwargs, cube):
+ """Update ``weights`` keyword argument in-place.
+
+ Args:
+
+ * kwargs (dict):
+ Keyword arguments that will be updated in-place if a `weights`
+ keyword is present which is not ``None``.
+ * cube (Cube):
+ Input cube for aggregation. If weights is given as :obj:`str`, try
+ to extract a cell measure with the corresponding name from this
+ cube. Otherwise, this argument is ignored.
+
+ """
+ if kwargs.get("weights") is not None:
+ kwargs["weights"] = cls(kwargs["weights"], cube)
+
+
def create_weighted_aggregator_fn(aggregator_fn, axis, **kwargs):
- """Return an aggregator function that can explicitely handle weights.
+ """Return an aggregator function that can explicitly handle weights.
Args:
@@ -1399,7 +1513,7 @@ def _weighted_quantile_1D(data, weights, quantiles, **kwargs):
array or float. Calculated quantile values (set to np.nan wherever sum
of weights is zero or masked)
"""
- # Return np.nan if no useable points found
+ # Return np.nan if no usable points found
if np.isclose(weights.sum(), 0.0) or ma.is_masked(weights.sum()):
return np.resize(np.array(np.nan), len(quantiles))
# Sort the data
@@ -1536,7 +1650,7 @@ def _proportion(array, function, axis, **kwargs):
# Otherwise, it is possible for numpy to return a masked array that has
# a dtype for its data that is different to the dtype of the fill-value,
# which can cause issues outside this function.
- # Reference - tests/unit/analyis/test_PROPORTION.py Test_masked.test_ma
+ # Reference - tests/unit/analysis/test_PROPORTION.py Test_masked.test_ma
numerator = _count(array, axis=axis, function=function, **kwargs)
result = ma.asarray(numerator / total_non_masked)
@@ -1584,27 +1698,19 @@ def _lazy_max_run(array, axis=-1, **kwargs):
def _rms(array, axis, **kwargs):
- # XXX due to the current limitations in `da.average` (see below), maintain
- # an explicit non-lazy aggregation function for now.
- # Note: retaining this function also means that if weights are passed to
- # the lazy aggregator, the aggregation will fall back to using this
- # non-lazy aggregator.
- rval = np.sqrt(ma.average(np.square(array), axis=axis, **kwargs))
- if not ma.isMaskedArray(array):
- rval = np.asarray(rval)
+ rval = np.sqrt(ma.average(array**2, axis=axis, **kwargs))
+
return rval
-@_build_dask_mdtol_function
def _lazy_rms(array, axis, **kwargs):
- # XXX This should use `da.average` and not `da.mean`, as does the above.
- # However `da.average` current doesn't handle masked weights correctly
- # (see https://github.com/dask/dask/issues/3846).
- # To work around this we use da.mean, which doesn't support weights at
- # all. Thus trying to use this aggregator with weights will currently
- # raise an error in dask due to the unexpected keyword `weights`,
- # rather than silently returning the wrong answer.
- return da.sqrt(da.mean(array**2, axis=axis, **kwargs))
+ # Note that, since we specifically need the ma version of average to handle
+ # weights correctly with masked data, we cannot rely on NEP13/18 and need
+ # to implement a separate lazy RMS function.
+
+ rval = da.sqrt(da.ma.average(array**2, axis=axis, **kwargs))
+
+ return rval
def _sum(array, **kwargs):
@@ -1639,6 +1745,18 @@ def _sum(array, **kwargs):
return rvalue
+def _sum_units_func(units, **kwargs):
+ """Multiply original units with weight units if possible."""
+ weights = kwargs.get("weights")
+ if weights is None: # no weights given or weights are None
+ result = units
+ elif hasattr(weights, "units"): # weights are _Weights
+ result = units * weights.units
+ else: # weights are regular np.ndarrays
+ result = units
+ return result
+
+
def _peak(array, **kwargs):
def column_segments(column):
nan_indices = np.where(np.isnan(column))[0]
@@ -1754,7 +1872,7 @@ def interp_order(length):
COUNT = Aggregator(
"count",
_count,
- units_func=lambda units: 1,
+ units_func=lambda units, **kwargs: 1,
lazy_func=_build_dask_mdtol_function(_count),
)
"""
@@ -1786,7 +1904,7 @@ def interp_order(length):
MAX_RUN = Aggregator(
None,
iris._lazy_data.non_lazy(_lazy_max_run),
- units_func=lambda units: 1,
+ units_func=lambda units, **kwargs: 1,
lazy_func=_build_dask_mdtol_function(_lazy_max_run),
)
"""
@@ -2030,7 +2148,11 @@ def interp_order(length):
"""
-PROPORTION = Aggregator("proportion", _proportion, units_func=lambda units: 1)
+PROPORTION = Aggregator(
+ "proportion",
+ _proportion,
+ units_func=lambda units, **kwargs: 1,
+)
"""
An :class:`~iris.analysis.Aggregator` instance that calculates the
proportion, as a fraction, of :class:`~iris.cube.Cube` data occurrences
@@ -2072,14 +2194,16 @@ def interp_order(length):
the root mean square over a :class:`~iris.cube.Cube`, as computed by
((x0**2 + x1**2 + ... + xN-1**2) / N) ** 0.5.
-Additional kwargs associated with the use of this aggregator:
+Parameters
+----------
-* weights (float ndarray):
+weights : array-like, optional
Weights matching the shape of the cube or the length of the window for
rolling window operations. The weights are applied to the squares when
taking the mean.
-**For example**:
+Example
+-------
To compute the zonal root mean square over the *longitude* axis of a cube::
@@ -2129,6 +2253,7 @@ def interp_order(length):
SUM = WeightedAggregator(
"sum",
_sum,
+ units_func=_sum_units_func,
lazy_func=_build_dask_mdtol_function(_sum),
)
"""
@@ -2166,7 +2291,7 @@ def interp_order(length):
VARIANCE = Aggregator(
"variance",
ma.var,
- units_func=lambda units: units * units,
+ units_func=lambda units, **kwargs: units * units,
lazy_func=_build_dask_mdtol_function(da.var),
ddof=1,
)
@@ -2808,7 +2933,7 @@ def __init__(self, mdtol=1):
Both sourge and target cubes must have an XY grid defined by
separate X and Y dimensions with dimension coordinates.
All of the XY dimension coordinates must also be bounded, and have
- the same cooordinate system.
+ the same coordinate system.
"""
if not (0 <= mdtol <= 1):
diff --git a/lib/iris/analysis/_area_weighted.py b/lib/iris/analysis/_area_weighted.py
index edbfd41ef9..3b728e9a43 100644
--- a/lib/iris/analysis/_area_weighted.py
+++ b/lib/iris/analysis/_area_weighted.py
@@ -853,7 +853,7 @@ def _calculate_regrid_area_weighted_weights(
cached_x_bounds = []
cached_x_indices = []
max_x_indices = 0
- for (x_0, x_1) in grid_x_bounds:
+ for x_0, x_1 in grid_x_bounds:
if grid_x_decreasing:
x_0, x_1 = x_1, x_0
x_bounds, x_indices = _cropped_bounds(src_x_bounds, x_0, x_1)
diff --git a/lib/iris/analysis/_interpolation.py b/lib/iris/analysis/_interpolation.py
index 2a7dfa6e62..f5e89a9e51 100644
--- a/lib/iris/analysis/_interpolation.py
+++ b/lib/iris/analysis/_interpolation.py
@@ -268,7 +268,7 @@ def _account_for_circular(self, points, data):
"""
from iris.analysis.cartography import wrap_lons
- for (circular, modulus, index, dim, offset) in self._circulars:
+ for circular, modulus, index, dim, offset in self._circulars:
if modulus:
# Map all the requested values into the range of the source
# data (centred over the centre of the source data to allow
diff --git a/lib/iris/analysis/_scipy_interpolate.py b/lib/iris/analysis/_scipy_interpolate.py
index fc64249729..bfa070c7c7 100644
--- a/lib/iris/analysis/_scipy_interpolate.py
+++ b/lib/iris/analysis/_scipy_interpolate.py
@@ -225,7 +225,6 @@ def compute_interp_weights(self, xi, method=None):
prepared = (xi_shape, method) + self._find_indices(xi.T)
if method == "linear":
-
xi_shape, method, indices, norm_distances, out_of_bounds = prepared
# Allocate arrays for describing the sparse matrix.
diff --git a/lib/iris/analysis/calculus.py b/lib/iris/analysis/calculus.py
index c530dbd216..75b7d86406 100644
--- a/lib/iris/analysis/calculus.py
+++ b/lib/iris/analysis/calculus.py
@@ -594,7 +594,6 @@ def curl(i_cube, j_cube, k_cube=None):
horiz_cs, (iris.coord_systems.GeogCS, iris.coord_systems.RotatedGeogCS)
)
if not spherical_coords:
-
# TODO Implement some mechanism for conforming to a common grid
dj_dx = _curl_differentiate(j_cube, x_coord)
prototype_diff = dj_dx
diff --git a/lib/iris/analysis/trajectory.py b/lib/iris/analysis/trajectory.py
index c21d71d48c..24f7a9dede 100644
--- a/lib/iris/analysis/trajectory.py
+++ b/lib/iris/analysis/trajectory.py
@@ -85,7 +85,6 @@ def __init__(self, waypoints, sample_count=10):
cur_seg = segments[cur_seg_i]
len_accum = cur_seg.length
for p in range(self.sample_count):
-
# calculate the sample position along our total length
sample_at_len = p * sample_step
diff --git a/lib/iris/common/metadata.py b/lib/iris/common/metadata.py
index 8ec39bb4b1..cb3149fe58 100644
--- a/lib/iris/common/metadata.py
+++ b/lib/iris/common/metadata.py
@@ -969,6 +969,7 @@ def _combine_lenient(self, other):
A list of combined metadata member values.
"""
+
# Perform "strict" combination for "coord_system" and "climatological".
def func(field):
left = getattr(self, field)
@@ -1024,6 +1025,7 @@ def _difference_lenient(self, other):
A list of difference metadata member values.
"""
+
# Perform "strict" difference for "coord_system" and "climatological".
def func(field):
left = getattr(self, field)
diff --git a/lib/iris/coord_categorisation.py b/lib/iris/coord_categorisation.py
index 72019b4b87..698b4828f1 100644
--- a/lib/iris/coord_categorisation.py
+++ b/lib/iris/coord_categorisation.py
@@ -90,6 +90,7 @@ def vectorised_fn(*args):
# coordinates only
#
+
# Private "helper" function
def _pt_date(coord, time):
"""
diff --git a/lib/iris/coord_systems.py b/lib/iris/coord_systems.py
index 802571925e..edf0c1871b 100644
--- a/lib/iris/coord_systems.py
+++ b/lib/iris/coord_systems.py
@@ -478,7 +478,6 @@ def datum(self, value):
@classmethod
def from_datum(cls, datum, longitude_of_prime_meridian=None):
-
crs = super().__new__(cls)
crs._semi_major_axis = None
@@ -949,7 +948,6 @@ def __init__(
false_northing=None,
ellipsoid=None,
):
-
"""
Constructs a Geostationary coord system.
diff --git a/lib/iris/coords.py b/lib/iris/coords.py
index cfcdcd92a0..91bb786ae8 100644
--- a/lib/iris/coords.py
+++ b/lib/iris/coords.py
@@ -2846,7 +2846,6 @@ def _new_bounds_requirements(self, bounds):
n_bounds = bounds.shape[-1]
n_points = bounds.shape[0]
if n_points > 1:
-
directions = set()
for b_index in range(n_bounds):
monotonic, direction = iris.util.monotonic(
diff --git a/lib/iris/cube.py b/lib/iris/cube.py
index be01bc9e5d..fcd7b5b828 100644
--- a/lib/iris/cube.py
+++ b/lib/iris/cube.py
@@ -28,6 +28,7 @@
import iris._lazy_data as _lazy
import iris._merge
import iris.analysis
+from iris.analysis import _Weights
from iris.analysis.cartography import wrap_lons
import iris.analysis.maths
import iris.aux_factory
@@ -884,7 +885,8 @@ def __init__(
This object defines the shape of the cube and the phenomenon
value in each cell.
- ``data`` can be a dask array, a NumPy array, a NumPy array
+ ``data`` can be a :class:`dask.array.Array`, a
+ :class:`numpy.ndarray`, a NumPy array
subclass (such as :class:`numpy.ma.MaskedArray`), or
array_like (as described in :func:`numpy.asarray`).
@@ -2687,7 +2689,6 @@ def subset(self, coord):
coord_to_extract in self.aux_coords
and len(coord_to_extract.points) == 1
):
-
# Default to returning None
result = None
@@ -3721,9 +3722,15 @@ def collapsed(self, coords, aggregator, **kwargs):
sum :data:`~iris.analysis.SUM`.
Weighted aggregations support an optional *weights* keyword argument.
- If set, this should be supplied as an array of weights whose shape
- matches the cube. Values for latitude-longitude area weights may be
- calculated using :func:`iris.analysis.cartography.area_weights`.
+ If set, this can be supplied as an array, cube, or (names of)
+ :meth:`~iris.cube.Cube.coords`, :meth:`~iris.cube.Cube.cell_measures`,
+ or :meth:`~iris.cube.Cube.ancillary_variables`. In all cases, the
+ weights should be 1d (for collapsing over a 1d coordinate) or match the
+ shape of the cube. When weights are not given as arrays, units are
+ correctly handled for weighted sums, i.e., the original unit of the
+ cube is multiplied by the units of the weights. Values for
+ latitude-longitude area weights may be calculated using
+ :func:`iris.analysis.cartography.area_weights`.
Some Iris aggregators support "lazy" evaluation, meaning that
cubes resulting from this method may represent data arrays which are
@@ -3802,6 +3809,10 @@ def collapsed(self, coords, aggregator, **kwargs):
cube.collapsed(['latitude', 'longitude'],
iris.analysis.VARIANCE)
"""
+ # Update weights kwargs (if necessary) to handle different types of
+ # weights
+ _Weights.update_kwargs(kwargs, self)
+
# Convert any coordinate names to coordinates
coords = self._as_list_of_coords(coords)
@@ -3970,10 +3981,14 @@ def aggregated_by(
also be supplied. These include :data:`~iris.analysis.MEAN` and
:data:`~iris.analysis.SUM`.
- Weighted aggregations support an optional *weights* keyword argument. If
- set, this should be supplied as an array of weights whose shape matches
- the cube or as 1D array whose length matches the dimension over which is
- aggregated.
+ Weighted aggregations support an optional *weights* keyword argument.
+ If set, this can be supplied as an array, cube, or (names of)
+ :meth:`~iris.cube.Cube.coords`, :meth:`~iris.cube.Cube.cell_measures`,
+ or :meth:`~iris.cube.Cube.ancillary_variables`. In all cases, the
+ weights should be 1d or match the shape of the cube. When weights are
+ not given as arrays, units are correctly handled for weighted sums,
+ i.e., the original unit of the cube is multiplied by the units of the
+ weights.
Parameters
----------
@@ -4032,6 +4047,10 @@ def aggregated_by(
STASH m01s00i024
"""
+ # Update weights kwargs (if necessary) to handle different types of
+ # weights
+ _Weights.update_kwargs(kwargs, self)
+
groupby_coords = []
dimension_to_groupby = None
@@ -4070,10 +4089,16 @@ def aggregated_by(
f"that is aggregated, got {len(weights):d}, expected "
f"{self.shape[dimension_to_groupby]:d}"
)
- weights = iris.util.broadcast_to_shape(
- weights,
- self.shape,
- (dimension_to_groupby,),
+
+ # iris.util.broadcast_to_shape does not preserve _Weights type
+ weights = _Weights(
+ iris.util.broadcast_to_shape(
+ weights,
+ self.shape,
+ (dimension_to_groupby,),
+ ),
+ self,
+ units=weights.units,
)
if weights.shape != self.shape:
raise ValueError(
@@ -4289,8 +4314,11 @@ def rolling_window(self, coord, aggregator, window, **kwargs):
* kwargs:
Aggregator and aggregation function keyword arguments. The weights
- argument to the aggregator, if any, should be a 1d array with the
- same length as the chosen window.
+ argument to the aggregator, if any, should be a 1d array, cube, or
+ (names of) :meth:`~iris.cube.Cube.coords`,
+ :meth:`~iris.cube.Cube.cell_measures`, or
+ :meth:`~iris.cube.Cube.ancillary_variables` with the same length as
+ the chosen window.
Returns:
:class:`iris.cube.Cube`.
@@ -4358,6 +4386,10 @@ def rolling_window(self, coord, aggregator, window, **kwargs):
possible windows of size 3 from the original cube.
"""
+ # Update weights kwargs (if necessary) to handle different types of
+ # weights
+ _Weights.update_kwargs(kwargs, self)
+
coord = self._as_list_of_coords(coord)[0]
if getattr(coord, "circular", False):
@@ -4459,8 +4491,14 @@ def rolling_window(self, coord, aggregator, window, **kwargs):
"as the window."
)
kwargs = dict(kwargs)
- kwargs["weights"] = iris.util.broadcast_to_shape(
- weights, rolling_window_data.shape, (dimension + 1,)
+
+ # iris.util.broadcast_to_shape does not preserve _Weights type
+ kwargs["weights"] = _Weights(
+ iris.util.broadcast_to_shape(
+ weights, rolling_window_data.shape, (dimension + 1,)
+ ),
+ self,
+ units=weights.units,
)
data_result = aggregator.aggregate(
rolling_window_data, axis=dimension + 1, **kwargs
diff --git a/lib/iris/experimental/ugrid/load.py b/lib/iris/experimental/ugrid/load.py
index a522d91313..cfa3935991 100644
--- a/lib/iris/experimental/ugrid/load.py
+++ b/lib/iris/experimental/ugrid/load.py
@@ -209,7 +209,8 @@ def load_meshes(uris, var_name=None):
result = {}
for source in valid_sources:
- meshes_dict = _meshes_from_cf(CFUGridReader(source))
+ with CFUGridReader(source) as cf_reader:
+ meshes_dict = _meshes_from_cf(cf_reader)
meshes = list(meshes_dict.values())
if var_name is not None:
meshes = list(filter(lambda m: m.var_name == var_name, meshes))
diff --git a/lib/iris/experimental/ugrid/mesh.py b/lib/iris/experimental/ugrid/mesh.py
index 4fd09175af..0d566da73f 100644
--- a/lib/iris/experimental/ugrid/mesh.py
+++ b/lib/iris/experimental/ugrid/mesh.py
@@ -131,7 +131,7 @@ def __init__(
Args:
- * indices (numpy.ndarray or numpy.ma.core.MaskedArray or dask.array.Array):
+ * indices (:class:`numpy.ndarray` or :class:`numpy.ma.core.MaskedArray` or :class:`dask.array.Array`):
2D array giving the topological connection relationship between
:attr:`location` elements and :attr:`connected` elements.
The :attr:`location_axis` dimension indexes over the
@@ -501,7 +501,7 @@ def core_indices(self):
NumPy array or a Dask array.
Returns:
- numpy.ndarray or numpy.ma.core.MaskedArray or dask.array.Array
+ :class:`numpy.ndarray` or :class:`numpy.ma.core.MaskedArray` or :class:`dask.array.Array`
"""
return super()._core_values()
@@ -3127,9 +3127,7 @@ def _construct_access_arrays(self):
flat_inds_safe = al.where(missing_inds, 0, flat_inds_nomask)
# Here's the core indexing operation.
# The comma applies all inds-array values to the *first* dimension.
- bounds = node_points[
- flat_inds_safe,
- ]
+ bounds = node_points[flat_inds_safe,]
# Fix 'missing' locations, and restore the proper shape.
bounds = al.ma.masked_array(bounds, missing_inds)
bounds = bounds.reshape(indices.shape)
diff --git a/lib/iris/experimental/ugrid/metadata.py b/lib/iris/experimental/ugrid/metadata.py
index ae0b787908..44bbe04fe9 100644
--- a/lib/iris/experimental/ugrid/metadata.py
+++ b/lib/iris/experimental/ugrid/metadata.py
@@ -53,6 +53,7 @@ def _combine_lenient(self, other):
A list of combined metadata member values.
"""
+
# Perform "strict" combination for "cf_role", "start_index", "location_axis".
def func(field):
left = getattr(self, field)
@@ -113,6 +114,7 @@ def _difference_lenient(self, other):
A list of difference metadata member values.
"""
+
# Perform "strict" difference for "cf_role", "start_index", "location_axis".
def func(field):
left = getattr(self, field)
@@ -233,6 +235,7 @@ def _difference_lenient(self, other):
A list of difference metadata member values.
"""
+
# Perform "strict" difference for "topology_dimension",
# "node_dimension", "edge_dimension" and "face_dimension".
def func(field):
@@ -297,6 +300,7 @@ def _combine_lenient(self, other):
A list of combined metadata member values.
"""
+
# It is actually "strict" : return None except where members are equal.
def func(field):
left = getattr(self, field)
@@ -352,6 +356,7 @@ def _difference_lenient(self, other):
A list of different metadata member values.
"""
+
# Perform "strict" difference for location / axis.
def func(field):
left = getattr(self, field)
diff --git a/lib/iris/fileformats/abf.py b/lib/iris/fileformats/abf.py
index 5c70c5acf2..4dcd5ce6aa 100644
--- a/lib/iris/fileformats/abf.py
+++ b/lib/iris/fileformats/abf.py
@@ -219,7 +219,6 @@ def load_cubes(filespecs, callback=None):
for filespec in filespecs:
for filename in glob.glob(filespec):
-
field = ABFField(filename)
cube = field.to_cube()
diff --git a/lib/iris/fileformats/cf.py b/lib/iris/fileformats/cf.py
index a3a23dc323..a21e1d975f 100644
--- a/lib/iris/fileformats/cf.py
+++ b/lib/iris/fileformats/cf.py
@@ -20,10 +20,10 @@
import re
import warnings
-import netCDF4
import numpy as np
import numpy.ma as ma
+from iris.fileformats.netcdf import _thread_safe_nc
import iris.util
#
@@ -1050,7 +1050,9 @@ def __init__(self, filename, warn=False, monotonic=False):
#: Collection of CF-netCDF variables associated with this netCDF file
self.cf_group = self.CFGroup()
- self._dataset = netCDF4.Dataset(self._filename, mode="r")
+ self._dataset = _thread_safe_nc.DatasetWrapper(
+ self._filename, mode="r"
+ )
# Issue load optimisation warning.
if warn and self._dataset.file_format in [
@@ -1068,6 +1070,19 @@ def __init__(self, filename, warn=False, monotonic=False):
self._build_cf_groups()
self._reset()
+ def __enter__(self):
+ # Enable use as a context manager
+ # N.B. this **guarantees* closure of the file, when the context is exited.
+ # Note: ideally, the class would not do so much work in the __init__ call, and
+ # would do all that here, after acquiring necessary permissions/locks.
+ # But for legacy reasons, we can't do that. So **effectively**, the context
+ # (in terms of access control) alreday started, when we created the object.
+ return self
+
+ def __exit__(self, exc_type, exc_value, traceback):
+ # When used as a context-manager, **always** close the file on exit.
+ self._close()
+
@property
def filename(self):
"""The file that the CFReader is reading."""
@@ -1294,10 +1309,15 @@ def _reset(self):
for nc_var_name in self._dataset.variables.keys():
self.cf_group[nc_var_name].cf_attrs_reset()
- def __del__(self):
+ def _close(self):
# Explicitly close dataset to prevent file remaining open.
if self._dataset is not None:
self._dataset.close()
+ self._dataset = None
+
+ def __del__(self):
+ # Be sure to close dataset when CFReader is destroyed / garbage-collected.
+ self._close()
def _getncattr(dataset, attr, default=None):
diff --git a/lib/iris/fileformats/name_loaders.py b/lib/iris/fileformats/name_loaders.py
index d15a3717d0..b9b64a343e 100644
--- a/lib/iris/fileformats/name_loaders.py
+++ b/lib/iris/fileformats/name_loaders.py
@@ -994,7 +994,6 @@ def load_NAMEIII_version2(filename):
# using the next() method. This will come in handy as we wish to
# progress through the file line by line.
with open(filename, "r") as file_handle:
-
# define a dictionary to hold the header metadata about this file
header = read_header(file_handle)
@@ -1005,7 +1004,6 @@ def load_NAMEIII_version2(filename):
column_headings = {}
datacol1 = header["Number of preliminary cols"]
for line in file_handle:
-
data = [col.strip() for col in line.split(",")][:-1]
# If first column is not zero we have reached the end
diff --git a/lib/iris/fileformats/netcdf/_thread_safe_nc.py b/lib/iris/fileformats/netcdf/_thread_safe_nc.py
new file mode 100644
index 0000000000..decca1535f
--- /dev/null
+++ b/lib/iris/fileformats/netcdf/_thread_safe_nc.py
@@ -0,0 +1,342 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""
+Module to ensure all calls to the netCDF4 library are thread-safe.
+
+Intention is that no other Iris module should import the netCDF4 module.
+
+"""
+from abc import ABC
+from threading import Lock
+import typing
+
+import netCDF4
+import numpy as np
+
+_GLOBAL_NETCDF4_LOCK = Lock()
+
+# Doesn't need thread protection, but this allows all netCDF4 refs to be
+# replaced with thread_safe refs.
+default_fillvals = netCDF4.default_fillvals
+
+
+class _ThreadSafeWrapper(ABC):
+ """
+ Contains a netCDF4 class instance, ensuring wrapping all API calls within _GLOBAL_NETCDF4_LOCK.
+
+ Designed to 'gate keep' all the instance's API calls, but allowing the
+ same API as if working directly with the instance itself.
+
+ Using a contained object instead of inheritance, as we cannot successfully
+ subclass or monkeypatch netCDF4 classes, because they are only wrappers for
+ the C-layer.
+ """
+
+ CONTAINED_CLASS = NotImplemented
+
+ # Allows easy type checking, avoiding difficulties with isinstance and mocking.
+ THREAD_SAFE_FLAG = True
+
+ @classmethod
+ def _from_existing(cls, instance):
+ """Pass an existing instance to __init__, where it is contained."""
+ assert isinstance(instance, cls.CONTAINED_CLASS)
+ return cls(instance)
+
+ def __init__(self, *args, **kwargs):
+ """Contain an existing instance, or generate a new one from arguments."""
+ if isinstance(args[0], self.CONTAINED_CLASS):
+ instance = args[0]
+ else:
+ with _GLOBAL_NETCDF4_LOCK:
+ instance = self.CONTAINED_CLASS(*args, **kwargs)
+
+ self._contained_instance = instance
+
+ def __getattr__(self, item):
+ if item == "_contained_instance":
+ # Special behaviour when accessing the _contained_instance itself.
+ return object.__getattribute__(self, item)
+ else:
+ with _GLOBAL_NETCDF4_LOCK:
+ return getattr(self._contained_instance, item)
+
+ def __setattr__(self, key, value):
+ if key == "_contained_instance":
+ # Special behaviour when accessing the _contained_instance itself.
+ object.__setattr__(self, key, value)
+ else:
+ with _GLOBAL_NETCDF4_LOCK:
+ return setattr(self._contained_instance, key, value)
+
+ def __getitem__(self, item):
+ with _GLOBAL_NETCDF4_LOCK:
+ return self._contained_instance.__getitem__(item)
+
+ def __setitem__(self, key, value):
+ with _GLOBAL_NETCDF4_LOCK:
+ return self._contained_instance.__setitem__(key, value)
+
+
+class DimensionWrapper(_ThreadSafeWrapper):
+ """
+ Accessor for a netCDF4.Dimension, always acquiring _GLOBAL_NETCDF4_LOCK.
+
+ All API calls should be identical to those for netCDF4.Dimension.
+ """
+
+ CONTAINED_CLASS = netCDF4.Dimension
+
+
+class VariableWrapper(_ThreadSafeWrapper):
+ """
+ Accessor for a netCDF4.Variable, always acquiring _GLOBAL_NETCDF4_LOCK.
+
+ All API calls should be identical to those for netCDF4.Variable.
+ """
+
+ CONTAINED_CLASS = netCDF4.Variable
+
+ def setncattr(self, *args, **kwargs) -> None:
+ """
+ Calls netCDF4.Variable.setncattr within _GLOBAL_NETCDF4_LOCK.
+
+ Only defined explicitly in order to get some mocks to work.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ return self._contained_instance.setncattr(*args, **kwargs)
+
+ @property
+ def dimensions(self) -> typing.List[str]:
+ """
+ Calls netCDF4.Variable.dimensions within _GLOBAL_NETCDF4_LOCK.
+
+ Only defined explicitly in order to get some mocks to work.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ # Return value is a list of strings so no need for
+ # DimensionWrapper, unlike self.get_dims().
+ return self._contained_instance.dimensions
+
+ # All Variable API that returns Dimension(s) is wrapped to instead return
+ # DimensionWrapper(s).
+
+ def get_dims(self, *args, **kwargs) -> typing.Tuple[DimensionWrapper]:
+ """
+ Calls netCDF4.Variable.get_dims() within _GLOBAL_NETCDF4_LOCK, returning DimensionWrappers.
+
+ The original returned netCDF4.Dimensions are simply replaced with their
+ respective DimensionWrappers, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ dimensions_ = list(
+ self._contained_instance.get_dims(*args, **kwargs)
+ )
+ return tuple([DimensionWrapper._from_existing(d) for d in dimensions_])
+
+
+class GroupWrapper(_ThreadSafeWrapper):
+ """
+ Accessor for a netCDF4.Group, always acquiring _GLOBAL_NETCDF4_LOCK.
+
+ All API calls should be identical to those for netCDF4.Group.
+ """
+
+ CONTAINED_CLASS = netCDF4.Group
+
+ # All Group API that returns Dimension(s) is wrapped to instead return
+ # DimensionWrapper(s).
+
+ @property
+ def dimensions(self) -> typing.Dict[str, DimensionWrapper]:
+ """
+ Calls dimensions of netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning DimensionWrappers.
+
+ The original returned netCDF4.Dimensions are simply replaced with their
+ respective DimensionWrappers, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ dimensions_ = self._contained_instance.dimensions
+ return {
+ k: DimensionWrapper._from_existing(v)
+ for k, v in dimensions_.items()
+ }
+
+ def createDimension(self, *args, **kwargs) -> DimensionWrapper:
+ """
+ Calls createDimension() from netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning DimensionWrapper.
+
+ The original returned netCDF4.Dimension is simply replaced with its
+ respective DimensionWrapper, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ new_dimension = self._contained_instance.createDimension(
+ *args, **kwargs
+ )
+ return DimensionWrapper._from_existing(new_dimension)
+
+ # All Group API that returns Variable(s) is wrapped to instead return
+ # VariableWrapper(s).
+
+ @property
+ def variables(self) -> typing.Dict[str, VariableWrapper]:
+ """
+ Calls variables of netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning VariableWrappers.
+
+ The original returned netCDF4.Variables are simply replaced with their
+ respective VariableWrappers, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ variables_ = self._contained_instance.variables
+ return {
+ k: VariableWrapper._from_existing(v) for k, v in variables_.items()
+ }
+
+ def createVariable(self, *args, **kwargs) -> VariableWrapper:
+ """
+ Calls createVariable() from netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning VariableWrapper.
+
+ The original returned netCDF4.Variable is simply replaced with its
+ respective VariableWrapper, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ new_variable = self._contained_instance.createVariable(
+ *args, **kwargs
+ )
+ return VariableWrapper._from_existing(new_variable)
+
+ def get_variables_by_attributes(
+ self, *args, **kwargs
+ ) -> typing.List[VariableWrapper]:
+ """
+ Calls get_variables_by_attributes() from netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning VariableWrappers.
+
+ The original returned netCDF4.Variables are simply replaced with their
+ respective VariableWrappers, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ variables_ = list(
+ self._contained_instance.get_variables_by_attributes(
+ *args, **kwargs
+ )
+ )
+ return [VariableWrapper._from_existing(v) for v in variables_]
+
+ # All Group API that returns Group(s) is wrapped to instead return
+ # GroupWrapper(s).
+
+ @property
+ def groups(self):
+ """
+ Calls groups of netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning GroupWrappers.
+
+ The original returned netCDF4.Groups are simply replaced with their
+ respective GroupWrappers, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ groups_ = self._contained_instance.groups
+ return {k: GroupWrapper._from_existing(v) for k, v in groups_.items()}
+
+ @property
+ def parent(self):
+ """
+ Calls parent of netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning a GroupWrapper.
+
+ The original returned netCDF4.Group is simply replaced with its
+ respective GroupWrapper, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ parent_ = self._contained_instance.parent
+ return GroupWrapper._from_existing(parent_)
+
+ def createGroup(self, *args, **kwargs):
+ """
+ Calls createGroup() from netCDF4.Group/Dataset within _GLOBAL_NETCDF4_LOCK, returning GroupWrapper.
+
+ The original returned netCDF4.Group is simply replaced with its
+ respective GroupWrapper, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ new_group = self._contained_instance.createGroup(*args, **kwargs)
+ return GroupWrapper._from_existing(new_group)
+
+
+class DatasetWrapper(GroupWrapper):
+ """
+ Accessor for a netCDF4.Dataset, always acquiring _GLOBAL_NETCDF4_LOCK.
+
+ All API calls should be identical to those for netCDF4.Dataset.
+ """
+
+ CONTAINED_CLASS = netCDF4.Dataset
+
+ @classmethod
+ def fromcdl(cls, *args, **kwargs):
+ """
+ Calls netCDF4.Dataset.fromcdl() within _GLOBAL_NETCDF4_LOCK, returning a DatasetWrapper.
+
+ The original returned netCDF4.Dataset is simply replaced with its
+ respective DatasetWrapper, ensuring that downstream calls are
+ also performed within _GLOBAL_NETCDF4_LOCK.
+ """
+ with _GLOBAL_NETCDF4_LOCK:
+ instance = cls.CONTAINED_CLASS.fromcdl(*args, **kwargs)
+ return cls._from_existing(instance)
+
+
+class NetCDFDataProxy:
+ """A reference to the data payload of a single NetCDF file variable."""
+
+ __slots__ = ("shape", "dtype", "path", "variable_name", "fill_value")
+
+ def __init__(self, shape, dtype, path, variable_name, fill_value):
+ self.shape = shape
+ self.dtype = dtype
+ self.path = path
+ self.variable_name = variable_name
+ self.fill_value = fill_value
+
+ @property
+ def ndim(self):
+ return len(self.shape)
+
+ def __getitem__(self, keys):
+ # Using a DatasetWrapper causes problems with invalid ID's and the
+ # netCDF4 library, presumably because __getitem__ gets called so many
+ # times by Dask. Use _GLOBAL_NETCDF4_LOCK directly instead.
+ with _GLOBAL_NETCDF4_LOCK:
+ dataset = netCDF4.Dataset(self.path)
+ try:
+ variable = dataset.variables[self.variable_name]
+ # Get the NetCDF variable data and slice.
+ var = variable[keys]
+ finally:
+ dataset.close()
+ return np.asanyarray(var)
+
+ def __repr__(self):
+ fmt = (
+ "<{self.__class__.__name__} shape={self.shape}"
+ " dtype={self.dtype!r} path={self.path!r}"
+ " variable_name={self.variable_name!r}>"
+ )
+ return fmt.format(self=self)
+
+ def __getstate__(self):
+ return {attr: getattr(self, attr) for attr in self.__slots__}
+
+ def __setstate__(self, state):
+ for key, value in state.items():
+ setattr(self, key, value)
diff --git a/lib/iris/fileformats/netcdf/loader.py b/lib/iris/fileformats/netcdf/loader.py
index 95f394c70d..8fcab61d17 100644
--- a/lib/iris/fileformats/netcdf/loader.py
+++ b/lib/iris/fileformats/netcdf/loader.py
@@ -15,7 +15,6 @@
"""
import warnings
-import netCDF4
import numpy as np
from iris._lazy_data import as_lazy_data
@@ -34,6 +33,7 @@
import iris.coords
import iris.exceptions
import iris.fileformats.cf
+from iris.fileformats.netcdf import _thread_safe_nc
from iris.fileformats.netcdf.saver import _CF_ATTRS
import iris.io
import iris.util
@@ -44,6 +44,10 @@
# Get the logger : shared logger for all in 'iris.fileformats.netcdf'.
from . import logger
+# An expected part of the public loader API, but includes thread safety
+# concerns so is housed in _thread_safe_nc.
+NetCDFDataProxy = _thread_safe_nc.NetCDFDataProxy
+
def _actions_engine():
# Return an 'actions engine', which provides a pyke-rules-like interface to
@@ -55,48 +59,6 @@ def _actions_engine():
return engine
-class NetCDFDataProxy:
- """A reference to the data payload of a single NetCDF file variable."""
-
- __slots__ = ("shape", "dtype", "path", "variable_name", "fill_value")
-
- def __init__(self, shape, dtype, path, variable_name, fill_value):
- self.shape = shape
- self.dtype = dtype
- self.path = path
- self.variable_name = variable_name
- self.fill_value = fill_value
-
- @property
- def ndim(self):
- return len(self.shape)
-
- def __getitem__(self, keys):
- dataset = netCDF4.Dataset(self.path)
- try:
- variable = dataset.variables[self.variable_name]
- # Get the NetCDF variable data and slice.
- var = variable[keys]
- finally:
- dataset.close()
- return np.asanyarray(var)
-
- def __repr__(self):
- fmt = (
- "<{self.__class__.__name__} shape={self.shape}"
- " dtype={self.dtype!r} path={self.path!r}"
- " variable_name={self.variable_name!r}>"
- )
- return fmt.format(self=self)
-
- def __getstate__(self):
- return {attr: getattr(self, attr) for attr in self.__slots__}
-
- def __setstate__(self, state):
- for key, value in state.items():
- setattr(self, key, value)
-
-
def _assert_case_specific_facts(engine, cf, cf_group):
# Initialise a data store for built cube elements.
# This is used to patch element attributes *not* setup by the actions
@@ -219,7 +181,7 @@ def _get_cf_var_data(cf_var, filename):
fill_value = getattr(
cf_var.cf_data,
"_FillValue",
- netCDF4.default_fillvals[cf_var.dtype.str[1:]],
+ _thread_safe_nc.default_fillvals[cf_var.dtype.str[1:]],
)
proxy = NetCDFDataProxy(
cf_var.shape, dtype, filename, cf_var.cf_name, fill_value
@@ -536,59 +498,62 @@ def load_cubes(filenames, callback=None, constraints=None):
# Ingest the netCDF file.
meshes = {}
if PARSE_UGRID_ON_LOAD:
- cf = CFUGridReader(filename)
- meshes = _meshes_from_cf(cf)
+ cf_reader_class = CFUGridReader
else:
- cf = iris.fileformats.cf.CFReader(filename)
+ cf_reader_class = iris.fileformats.cf.CFReader
- # Process each CF data variable.
- data_variables = list(cf.cf_group.data_variables.values()) + list(
- cf.cf_group.promoted.values()
- )
- for cf_var in data_variables:
- if var_callback and not var_callback(cf_var):
- # Deliver only selected results.
- continue
-
- # cf_var-specific mesh handling, if a mesh is present.
- # Build the mesh_coords *before* loading the cube - avoids
- # mesh-related attributes being picked up by
- # _add_unused_attributes().
- mesh_name = None
- mesh = None
- mesh_coords, mesh_dim = [], None
+ with cf_reader_class(filename) as cf:
if PARSE_UGRID_ON_LOAD:
- mesh_name = getattr(cf_var, "mesh", None)
- if mesh_name is not None:
+ meshes = _meshes_from_cf(cf)
+
+ # Process each CF data variable.
+ data_variables = list(cf.cf_group.data_variables.values()) + list(
+ cf.cf_group.promoted.values()
+ )
+ for cf_var in data_variables:
+ if var_callback and not var_callback(cf_var):
+ # Deliver only selected results.
+ continue
+
+ # cf_var-specific mesh handling, if a mesh is present.
+ # Build the mesh_coords *before* loading the cube - avoids
+ # mesh-related attributes being picked up by
+ # _add_unused_attributes().
+ mesh_name = None
+ mesh = None
+ mesh_coords, mesh_dim = [], None
+ if PARSE_UGRID_ON_LOAD:
+ mesh_name = getattr(cf_var, "mesh", None)
+ if mesh_name is not None:
+ try:
+ mesh = meshes[mesh_name]
+ except KeyError:
+ message = (
+ f"File does not contain mesh: '{mesh_name}' - "
+ f"referenced by variable: '{cf_var.cf_name}' ."
+ )
+ logger.debug(message)
+ if mesh is not None:
+ mesh_coords, mesh_dim = _build_mesh_coords(mesh, cf_var)
+
+ cube = _load_cube(engine, cf, cf_var, filename)
+
+ # Attach the mesh (if present) to the cube.
+ for mesh_coord in mesh_coords:
+ cube.add_aux_coord(mesh_coord, mesh_dim)
+
+ # Process any associated formula terms and attach
+ # the corresponding AuxCoordFactory.
try:
- mesh = meshes[mesh_name]
- except KeyError:
- message = (
- f"File does not contain mesh: '{mesh_name}' - "
- f"referenced by variable: '{cf_var.cf_name}' ."
- )
- logger.debug(message)
- if mesh is not None:
- mesh_coords, mesh_dim = _build_mesh_coords(mesh, cf_var)
-
- cube = _load_cube(engine, cf, cf_var, filename)
-
- # Attach the mesh (if present) to the cube.
- for mesh_coord in mesh_coords:
- cube.add_aux_coord(mesh_coord, mesh_dim)
-
- # Process any associated formula terms and attach
- # the corresponding AuxCoordFactory.
- try:
- _load_aux_factory(engine, cube)
- except ValueError as e:
- warnings.warn("{}".format(e))
-
- # Perform any user registered callback function.
- cube = run_callback(callback, cube, cf_var, filename)
-
- # Callback mechanism may return None, which must not be yielded
- if cube is None:
- continue
-
- yield cube
+ _load_aux_factory(engine, cube)
+ except ValueError as e:
+ warnings.warn("{}".format(e))
+
+ # Perform any user registered callback function.
+ cube = run_callback(callback, cube, cf_var, filename)
+
+ # Callback mechanism may return None, which must not be yielded
+ if cube is None:
+ continue
+
+ yield cube
diff --git a/lib/iris/fileformats/netcdf/saver.py b/lib/iris/fileformats/netcdf/saver.py
index 650c5e3338..f7f4864f9e 100644
--- a/lib/iris/fileformats/netcdf/saver.py
+++ b/lib/iris/fileformats/netcdf/saver.py
@@ -24,7 +24,6 @@
import cf_units
import dask.array as da
-import netCDF4
import numpy as np
import numpy.ma as ma
@@ -45,6 +44,7 @@
from iris.coords import AncillaryVariable, AuxCoord, CellMeasure, DimCoord
import iris.exceptions
import iris.fileformats.cf
+from iris.fileformats.netcdf import _thread_safe_nc
import iris.io
import iris.util
@@ -459,7 +459,10 @@ def _setncattr(variable, name, attribute):
Put the given attribute on the given netCDF4 Data type, casting
attributes as we go to bytes rather than unicode.
+ NOTE: variable needs to be a _thread_safe_nc._ThreadSafeWrapper subclass.
+
"""
+ assert hasattr(variable, "THREAD_SAFE_FLAG")
attribute = _bytes_if_ascii(attribute)
return variable.setncattr(name, attribute)
@@ -470,9 +473,12 @@ class _FillValueMaskCheckAndStoreTarget:
given value and whether it was masked, before passing the chunk to the
given target.
+ NOTE: target needs to be a _thread_safe_nc._ThreadSafeWrapper subclass.
+
"""
def __init__(self, target, fill_value=None):
+ assert hasattr(target, "THREAD_SAFE_FLAG")
self.target = target
self.fill_value = fill_value
self.contains_value = False
@@ -544,7 +550,7 @@ def __init__(self, filename, netcdf_format):
self._formula_terms_cache = {}
#: NetCDF dataset
try:
- self._dataset = netCDF4.Dataset(
+ self._dataset = _thread_safe_nc.DatasetWrapper(
filename, mode="w", format=netcdf_format
)
except RuntimeError:
@@ -1927,16 +1933,6 @@ def _create_generic_cf_array_var(
# Check if this is a dim-coord.
is_dimcoord = cube is not None and element in cube.dim_coords
- if isinstance(element, iris.coords.CellMeasure):
- # Disallow saving of *masked* cell measures.
- # NOTE: currently, this is the only functional difference in
- # variable creation between an ancillary and a cell measure.
- if iris.util.is_masked(data):
- # We can't save masked points properly, as we don't maintain
- # a fill_value. (Load will not record one, either).
- msg = "Cell measures with missing data are not supported."
- raise ValueError(msg)
-
if is_dimcoord:
# By definition of a CF-netCDF coordinate variable this
# coordinate must be 1-D and the name of the CF-netCDF variable
@@ -2331,7 +2327,13 @@ def _create_cf_data_variable(
dtype = data.dtype.newbyteorder("=")
def set_packing_ncattrs(cfvar):
- """Set netCDF packing attributes."""
+ """
+ Set netCDF packing attributes.
+
+ NOTE: cfvar needs to be a _thread_safe_nc._ThreadSafeWrapper subclass.
+
+ """
+ assert hasattr(cfvar, "THREAD_SAFE_FLAG")
if packing:
if scale_factor:
_setncattr(cfvar, "scale_factor", scale_factor)
@@ -2478,7 +2480,9 @@ def store(data, cf_var, fill_value):
if fill_value is not None:
fill_value_to_check = fill_value
else:
- fill_value_to_check = netCDF4.default_fillvals[dtype.str[1:]]
+ fill_value_to_check = _thread_safe_nc.default_fillvals[
+ dtype.str[1:]
+ ]
else:
fill_value_to_check = None
diff --git a/lib/iris/fileformats/pp.py b/lib/iris/fileformats/pp.py
index bc35acb3b3..cff088cf89 100644
--- a/lib/iris/fileformats/pp.py
+++ b/lib/iris/fileformats/pp.py
@@ -625,7 +625,7 @@ def __getstate__(self):
def __setstate__(self, state):
# Because we have __slots__, this is needed to support Pickle.load()
# (Use setattr, as there is no object dictionary.)
- for (key, value) in state:
+ for key, value in state:
setattr(self, key, value)
def __eq__(self, other):
@@ -2029,10 +2029,8 @@ def pp_filter(field):
res = True
if field.stash not in _STASH_ALLOW:
if pp_constraints.get("stash"):
-
res = False
for call_func in pp_constraints["stash"]:
-
if call_func(str(field.stash)):
res = True
break
diff --git a/lib/iris/fileformats/pp_load_rules.py b/lib/iris/fileformats/pp_load_rules.py
index ebccec47ee..11d03e978a 100644
--- a/lib/iris/fileformats/pp_load_rules.py
+++ b/lib/iris/fileformats/pp_load_rules.py
@@ -756,7 +756,6 @@ def date2year(t_in):
)
)
):
-
coords_and_dims.append(
_new_coord_and_dims(
do_vector,
diff --git a/lib/iris/fileformats/rules.py b/lib/iris/fileformats/rules.py
index 07ed5eb8ce..51940b7c4d 100644
--- a/lib/iris/fileformats/rules.py
+++ b/lib/iris/fileformats/rules.py
@@ -394,7 +394,7 @@ def _load_pairs_from_fields_and_filenames(
yield (cube, field)
regrid_cache = {}
- for (cube, factories, field) in results_needing_reference:
+ for cube, factories, field in results_needing_reference:
_resolve_factory_references(
cube, factories, concrete_reference_targets, regrid_cache
)
diff --git a/lib/iris/fileformats/um/_fast_load_structured_fields.py b/lib/iris/fileformats/um/_fast_load_structured_fields.py
index d193aa30ce..64b7f8e891 100644
--- a/lib/iris/fileformats/um/_fast_load_structured_fields.py
+++ b/lib/iris/fileformats/um/_fast_load_structured_fields.py
@@ -133,6 +133,7 @@ def element_arrays_and_dims(self):
def _field_vector_element_arrays(self):
"""Define the field components used in the structure analysis."""
+
# Define functions to make t1 and t2 values as date-time tuples.
# These depend on header version (PPField2 has no seconds values).
def t1_fn(fld):
diff --git a/lib/iris/io/format_picker.py b/lib/iris/io/format_picker.py
index edf448e95b..a8e333c566 100644
--- a/lib/iris/io/format_picker.py
+++ b/lib/iris/io/format_picker.py
@@ -134,8 +134,9 @@ def get_spec(self, basename, buffer_obj):
value = value[:50] + "..."
printable_values[key] = value
msg = (
- "No format specification could be found for the given buffer."
- " File element cache:\n {}".format(printable_values)
+ "No format specification could be found for the given buffer. "
+ "Perhaps a plugin is missing or has not been loaded. "
+ "File element cache:\n {}".format(printable_values)
)
raise ValueError(msg)
diff --git a/lib/iris/pandas.py b/lib/iris/pandas.py
index faa250285e..417b6b11de 100644
--- a/lib/iris/pandas.py
+++ b/lib/iris/pandas.py
@@ -238,7 +238,7 @@ def as_cubes(
A :class:`~pandas.DataFrame` using columns as a second data dimension will
need to be 'melted' before conversion. See the Examples for how.
- Dask ``DataFrame``\\s are not supported.
+ :class:`dask.dataframe.DataFrame`\\ s are not supported.
Examples
--------
@@ -686,7 +686,7 @@ def as_data_frame(
Notes
-----
- Dask ``DataFrame``\\s are not supported.
+ :class:`dask.dataframe.DataFrame`\\ s are not supported.
A :class:`~pandas.MultiIndex` :class:`~pandas.DataFrame` is returned by default.
Use the :meth:`~pandas.DataFrame.reset_index` to return a
diff --git a/lib/iris/plugins/README.md b/lib/iris/plugins/README.md
new file mode 100644
index 0000000000..e8dee1de2c
--- /dev/null
+++ b/lib/iris/plugins/README.md
@@ -0,0 +1,10 @@
+# Iris plugins
+
+`iris.plugins` is a [namespace package] allowing arbitrary plugins to be
+installed alongside Iris.
+
+See [the Iris documentation][plugins] for more information.
+
+
+[namespace package]: https://packaging.python.org/en/latest/guides/packaging-namespace-packages/
+[plugins]: https://scitools-iris.readthedocs.io/en/latest/community/plugins.html
diff --git a/lib/iris/quickplot.py b/lib/iris/quickplot.py
index 18ed2554a3..6006314265 100644
--- a/lib/iris/quickplot.py
+++ b/lib/iris/quickplot.py
@@ -45,7 +45,6 @@ def _title(cube_or_coord, with_units):
or units.is_no_unit()
or units == cf_units.Unit("1")
):
-
if _use_symbol(units):
units = units.symbol
elif units.is_time_reference():
diff --git a/lib/iris/tests/graphics/__init__.py b/lib/iris/tests/graphics/__init__.py
index a083de3934..544d989564 100755
--- a/lib/iris/tests/graphics/__init__.py
+++ b/lib/iris/tests/graphics/__init__.py
@@ -187,7 +187,6 @@ def check_graphic(test_id: str, results_dir: Union[str, Path]) -> None:
try:
def _create_missing(phash: str) -> None:
-
output_path = test_output_dir / (test_id + ".png")
print(f"Creating image file: {output_path}")
@@ -214,7 +213,6 @@ def _create_missing(phash: str) -> None:
phash = get_phash(buffer)
if test_id in repo:
-
expected = hex_to_hash(repo[test_id])
# Calculate hamming distance vector for the result hash.
diff --git a/lib/iris/tests/integration/netcdf/__init__.py b/lib/iris/tests/integration/netcdf/__init__.py
new file mode 100644
index 0000000000..f500b52520
--- /dev/null
+++ b/lib/iris/tests/integration/netcdf/__init__.py
@@ -0,0 +1,6 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""Integration tests for loading and saving netcdf files."""
diff --git a/lib/iris/tests/integration/netcdf/test_attributes.py b/lib/iris/tests/integration/netcdf/test_attributes.py
new file mode 100644
index 0000000000..a73d6c7d49
--- /dev/null
+++ b/lib/iris/tests/integration/netcdf/test_attributes.py
@@ -0,0 +1,119 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""Integration tests for attribute-related loading and saving netcdf files."""
+
+# Import iris.tests first so that some things can be initialised before
+# importing anything else.
+import iris.tests as tests # isort:skip
+
+from contextlib import contextmanager
+from unittest import mock
+
+import iris
+from iris.cube import Cube, CubeList
+from iris.fileformats.netcdf import CF_CONVENTIONS_VERSION
+
+
+class TestUmVersionAttribute(tests.IrisTest):
+ def test_single_saves_as_global(self):
+ cube = Cube(
+ [1.0],
+ standard_name="air_temperature",
+ units="K",
+ attributes={"um_version": "4.3"},
+ )
+ with self.temp_filename(".nc") as nc_path:
+ iris.save(cube, nc_path)
+ self.assertCDL(nc_path)
+
+ def test_multiple_same_saves_as_global(self):
+ cube_a = Cube(
+ [1.0],
+ standard_name="air_temperature",
+ units="K",
+ attributes={"um_version": "4.3"},
+ )
+ cube_b = Cube(
+ [1.0],
+ standard_name="air_pressure",
+ units="hPa",
+ attributes={"um_version": "4.3"},
+ )
+ with self.temp_filename(".nc") as nc_path:
+ iris.save(CubeList([cube_a, cube_b]), nc_path)
+ self.assertCDL(nc_path)
+
+ def test_multiple_different_saves_on_variables(self):
+ cube_a = Cube(
+ [1.0],
+ standard_name="air_temperature",
+ units="K",
+ attributes={"um_version": "4.3"},
+ )
+ cube_b = Cube(
+ [1.0],
+ standard_name="air_pressure",
+ units="hPa",
+ attributes={"um_version": "4.4"},
+ )
+ with self.temp_filename(".nc") as nc_path:
+ iris.save(CubeList([cube_a, cube_b]), nc_path)
+ self.assertCDL(nc_path)
+
+
+@contextmanager
+def _patch_site_configuration():
+ def cf_patch_conventions(conventions):
+ return ", ".join([conventions, "convention1, convention2"])
+
+ def update(config):
+ config["cf_profile"] = mock.Mock(name="cf_profile")
+ config["cf_patch"] = mock.Mock(name="cf_patch")
+ config["cf_patch_conventions"] = cf_patch_conventions
+
+ orig_site_config = iris.site_configuration.copy()
+ update(iris.site_configuration)
+ yield
+ iris.site_configuration = orig_site_config
+
+
+class TestConventionsAttributes(tests.IrisTest):
+ def test_patching_conventions_attribute(self):
+ # Ensure that user defined conventions are wiped and those which are
+ # saved patched through site_config can be loaded without an exception
+ # being raised.
+ cube = Cube(
+ [1.0],
+ standard_name="air_temperature",
+ units="K",
+ attributes={"Conventions": "some user defined conventions"},
+ )
+
+ # Patch the site configuration dictionary.
+ with _patch_site_configuration(), self.temp_filename(".nc") as nc_path:
+ iris.save(cube, nc_path)
+ res = iris.load_cube(nc_path)
+
+ self.assertEqual(
+ res.attributes["Conventions"],
+ "{}, {}, {}".format(
+ CF_CONVENTIONS_VERSION, "convention1", "convention2"
+ ),
+ )
+
+
+class TestStandardName(tests.IrisTest):
+ def test_standard_name_roundtrip(self):
+ standard_name = "air_temperature detection_minimum"
+ cube = iris.cube.Cube(1, standard_name=standard_name)
+ with self.temp_filename(suffix=".nc") as fout:
+ iris.save(cube, fout)
+ detection_limit_cube = iris.load_cube(fout)
+ self.assertEqual(detection_limit_cube.standard_name, standard_name)
+
+
+if __name__ == "__main__":
+ tests.main()
diff --git a/lib/iris/tests/integration/netcdf/test_aux_factories.py b/lib/iris/tests/integration/netcdf/test_aux_factories.py
new file mode 100644
index 0000000000..d89f275336
--- /dev/null
+++ b/lib/iris/tests/integration/netcdf/test_aux_factories.py
@@ -0,0 +1,160 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""Integration tests for aux-factory-related loading and saving netcdf files."""
+
+# Import iris.tests first so that some things can be initialised before
+# importing anything else.
+import iris.tests as tests # isort:skip
+
+import iris
+from iris.tests import stock as stock
+
+
+@tests.skip_data
+class TestAtmosphereSigma(tests.IrisTest):
+ def setUp(self):
+ # Modify stock cube so it is suitable to have a atmosphere sigma
+ # factory added to it.
+ cube = stock.realistic_4d_no_derived()
+ cube.coord("surface_altitude").rename("surface_air_pressure")
+ cube.coord("surface_air_pressure").units = "Pa"
+ cube.coord("sigma").units = "1"
+ ptop_coord = iris.coords.AuxCoord(1000.0, var_name="ptop", units="Pa")
+ cube.add_aux_coord(ptop_coord, ())
+ cube.remove_coord("level_height")
+ # Construct and add atmosphere sigma factory.
+ factory = iris.aux_factory.AtmosphereSigmaFactory(
+ cube.coord("ptop"),
+ cube.coord("sigma"),
+ cube.coord("surface_air_pressure"),
+ )
+ cube.add_aux_factory(factory)
+ self.cube = cube
+
+ def test_save(self):
+ with self.temp_filename(suffix=".nc") as filename:
+ iris.save(self.cube, filename)
+ self.assertCDL(filename)
+
+ def test_save_load_loop(self):
+ # Ensure that the AtmosphereSigmaFactory is automatically loaded
+ # when loading the file.
+ with self.temp_filename(suffix=".nc") as filename:
+ iris.save(self.cube, filename)
+ cube = iris.load_cube(filename, "air_potential_temperature")
+ assert cube.coords("air_pressure")
+
+
+@tests.skip_data
+class TestHybridPressure(tests.IrisTest):
+ def setUp(self):
+ # Modify stock cube so it is suitable to have a
+ # hybrid pressure factory added to it.
+ cube = stock.realistic_4d_no_derived()
+ cube.coord("surface_altitude").rename("surface_air_pressure")
+ cube.coord("surface_air_pressure").units = "Pa"
+ cube.coord("level_height").rename("level_pressure")
+ cube.coord("level_pressure").units = "Pa"
+ # Construct and add hybrid pressure factory.
+ factory = iris.aux_factory.HybridPressureFactory(
+ cube.coord("level_pressure"),
+ cube.coord("sigma"),
+ cube.coord("surface_air_pressure"),
+ )
+ cube.add_aux_factory(factory)
+ self.cube = cube
+
+ def test_save(self):
+ with self.temp_filename(suffix=".nc") as filename:
+ iris.save(self.cube, filename)
+ self.assertCDL(filename)
+
+ def test_save_load_loop(self):
+ # Tests an issue where the variable names in the formula
+ # terms changed to the standard_names instead of the variable names
+ # when loading a previously saved cube.
+ with self.temp_filename(suffix=".nc") as filename, self.temp_filename(
+ suffix=".nc"
+ ) as other_filename:
+ iris.save(self.cube, filename)
+ cube = iris.load_cube(filename, "air_potential_temperature")
+ iris.save(cube, other_filename)
+ other_cube = iris.load_cube(
+ other_filename, "air_potential_temperature"
+ )
+ self.assertEqual(cube, other_cube)
+
+
+@tests.skip_data
+class TestSaveMultipleAuxFactories(tests.IrisTest):
+ def test_hybrid_height_and_pressure(self):
+ cube = stock.realistic_4d()
+ cube.add_aux_coord(
+ iris.coords.DimCoord(
+ 1200.0, long_name="level_pressure", units="hPa"
+ )
+ )
+ cube.add_aux_coord(
+ iris.coords.DimCoord(0.5, long_name="other sigma", units="1")
+ )
+ cube.add_aux_coord(
+ iris.coords.DimCoord(
+ 1000.0, long_name="surface_air_pressure", units="hPa"
+ )
+ )
+ factory = iris.aux_factory.HybridPressureFactory(
+ cube.coord("level_pressure"),
+ cube.coord("other sigma"),
+ cube.coord("surface_air_pressure"),
+ )
+ cube.add_aux_factory(factory)
+ with self.temp_filename(suffix=".nc") as filename:
+ iris.save(cube, filename)
+ self.assertCDL(filename)
+
+ def test_shared_primary(self):
+ cube = stock.realistic_4d()
+ factory = iris.aux_factory.HybridHeightFactory(
+ cube.coord("level_height"),
+ cube.coord("sigma"),
+ cube.coord("surface_altitude"),
+ )
+ factory.rename("another altitude")
+ cube.add_aux_factory(factory)
+ with self.temp_filename(
+ suffix=".nc"
+ ) as filename, self.assertRaisesRegex(
+ ValueError, "multiple aux factories"
+ ):
+ iris.save(cube, filename)
+
+ def test_hybrid_height_cubes(self):
+ hh1 = stock.simple_4d_with_hybrid_height()
+ hh1.attributes["cube"] = "hh1"
+ hh2 = stock.simple_4d_with_hybrid_height()
+ hh2.attributes["cube"] = "hh2"
+ sa = hh2.coord("surface_altitude")
+ sa.points = sa.points * 10
+ with self.temp_filename(".nc") as fname:
+ iris.save([hh1, hh2], fname)
+ cubes = iris.load(fname, "air_temperature")
+ cubes = sorted(cubes, key=lambda cube: cube.attributes["cube"])
+ self.assertCML(cubes)
+
+ def test_hybrid_height_cubes_on_dimension_coordinate(self):
+ hh1 = stock.hybrid_height()
+ hh2 = stock.hybrid_height()
+ sa = hh2.coord("surface_altitude")
+ sa.points = sa.points * 10
+ emsg = "Unable to create dimensonless vertical coordinate."
+ with self.temp_filename(".nc") as fname, self.assertRaisesRegex(
+ ValueError, emsg
+ ):
+ iris.save([hh1, hh2], fname)
+
+
+if __name__ == "__main__":
+ tests.main()
diff --git a/lib/iris/tests/integration/netcdf/test_coord_systems.py b/lib/iris/tests/integration/netcdf/test_coord_systems.py
new file mode 100644
index 0000000000..8576f5ffe8
--- /dev/null
+++ b/lib/iris/tests/integration/netcdf/test_coord_systems.py
@@ -0,0 +1,281 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""Integration tests for coord-system-related loading and saving netcdf files."""
+
+# Import iris.tests first so that some things can be initialised before
+# importing anything else.
+import iris.tests as tests # isort:skip
+
+from os.path import join as path_join
+import shutil
+import tempfile
+
+import iris
+from iris.coords import DimCoord
+from iris.cube import Cube
+from iris.tests import stock as stock
+from iris.tests.stock.netcdf import ncgen_from_cdl
+from iris.tests.unit.fileformats.netcdf import test_load_cubes as tlc
+
+
+@tests.skip_data
+class TestCoordSystem(tests.IrisTest):
+ def setUp(self):
+ tlc.setUpModule()
+
+ def tearDown(self):
+ tlc.tearDownModule()
+
+ def test_load_laea_grid(self):
+ cube = iris.load_cube(
+ tests.get_data_path(
+ ("NetCDF", "lambert_azimuthal_equal_area", "euro_air_temp.nc")
+ )
+ )
+ self.assertCML(cube, ("netcdf", "netcdf_laea.cml"))
+
+ datum_cf_var_cdl = """
+ netcdf output {
+ dimensions:
+ y = 4 ;
+ x = 3 ;
+ variables:
+ float data(y, x) ;
+ data :standard_name = "toa_brightness_temperature" ;
+ data :units = "K" ;
+ data :grid_mapping = "mercator" ;
+ int mercator ;
+ mercator:grid_mapping_name = "mercator" ;
+ mercator:longitude_of_prime_meridian = 0. ;
+ mercator:earth_radius = 6378169. ;
+ mercator:horizontal_datum_name = "OSGB36" ;
+ float y(y) ;
+ y:axis = "Y" ;
+ y:units = "m" ;
+ y:standard_name = "projection_y_coordinate" ;
+ float x(x) ;
+ x:axis = "X" ;
+ x:units = "m" ;
+ x:standard_name = "projection_x_coordinate" ;
+
+ // global attributes:
+ :Conventions = "CF-1.7" ;
+ :standard_name_vocabulary = "CF Standard Name Table v27" ;
+
+ data:
+
+ data =
+ 0, 1, 2,
+ 3, 4, 5,
+ 6, 7, 8,
+ 9, 10, 11 ;
+
+ mercator = _ ;
+
+ y = 1, 2, 3, 5 ;
+
+ x = -6, -4, -2 ;
+
+ }
+ """
+
+ datum_wkt_cdl = """
+netcdf output5 {
+dimensions:
+ y = 4 ;
+ x = 3 ;
+variables:
+ float data(y, x) ;
+ data :standard_name = "toa_brightness_temperature" ;
+ data :units = "K" ;
+ data :grid_mapping = "mercator" ;
+ int mercator ;
+ mercator:grid_mapping_name = "mercator" ;
+ mercator:longitude_of_prime_meridian = 0. ;
+ mercator:earth_radius = 6378169. ;
+ mercator:longitude_of_projection_origin = 0. ;
+ mercator:false_easting = 0. ;
+ mercator:false_northing = 0. ;
+ mercator:scale_factor_at_projection_origin = 1. ;
+ mercator:crs_wkt = "PROJCRS[\\"unknown\\",BASEGEOGCRS[\\"unknown\\",DATUM[\\"OSGB36\\",ELLIPSOID[\\"unknown\\",6378169,0,LENGTHUNIT[\\"metre\\",1,ID[\\"EPSG\\",9001]]]],PRIMEM[\\"Greenwich\\",0,ANGLEUNIT[\\"degree\\",0.0174532925199433],ID[\\"EPSG\\",8901]]],CONVERSION[\\"unknown\\",METHOD[\\"Mercator (variant B)\\",ID[\\"EPSG\\",9805]],PARAMETER[\\"Latitude of 1st standard parallel\\",0,ANGLEUNIT[\\"degree\\",0.0174532925199433],ID[\\"EPSG\\",8823]],PARAMETER[\\"Longitude of natural origin\\",0,ANGLEUNIT[\\"degree\\",0.0174532925199433],ID[\\"EPSG\\",8802]],PARAMETER[\\"False easting\\",0,LENGTHUNIT[\\"metre\\",1],ID[\\"EPSG\\",8806]],PARAMETER[\\"False northing\\",0,LENGTHUNIT[\\"metre\\",1],ID[\\"EPSG\\",8807]]],CS[Cartesian,2],AXIS[\\"(E)\\",east,ORDER[1],LENGTHUNIT[\\"metre\\",1,ID[\\"EPSG\\",9001]]],AXIS[\\"(N)\\",north,ORDER[2],LENGTHUNIT[\\"metre\\",1,ID[\\"EPSG\\",9001]]]]" ;
+ float y(y) ;
+ y:axis = "Y" ;
+ y:units = "m" ;
+ y:standard_name = "projection_y_coordinate" ;
+ float x(x) ;
+ x:axis = "X" ;
+ x:units = "m" ;
+ x:standard_name = "projection_x_coordinate" ;
+
+// global attributes:
+ :standard_name_vocabulary = "CF Standard Name Table v27" ;
+ :Conventions = "CF-1.7" ;
+data:
+
+ data =
+ 0, 1, 2,
+ 3, 4, 5,
+ 6, 7, 8,
+ 9, 10, 11 ;
+
+ mercator = _ ;
+
+ y = 1, 2, 3, 5 ;
+
+ x = -6, -4, -2 ;
+}
+ """
+
+ def test_load_datum_wkt(self):
+ expected = "OSGB 1936"
+ nc_path = tlc.cdl_to_nc(self.datum_wkt_cdl)
+ with iris.FUTURE.context(datum_support=True):
+ cube = iris.load_cube(nc_path)
+ test_crs = cube.coord("projection_y_coordinate").coord_system
+ actual = str(test_crs.as_cartopy_crs().datum)
+ self.assertMultiLineEqual(expected, actual)
+
+ def test_no_load_datum_wkt(self):
+ nc_path = tlc.cdl_to_nc(self.datum_wkt_cdl)
+ with self.assertWarnsRegex(FutureWarning, "iris.FUTURE.datum_support"):
+ cube = iris.load_cube(nc_path)
+ test_crs = cube.coord("projection_y_coordinate").coord_system
+ actual = str(test_crs.as_cartopy_crs().datum)
+ self.assertMultiLineEqual(actual, "unknown")
+
+ def test_load_datum_cf_var(self):
+ expected = "OSGB 1936"
+ nc_path = tlc.cdl_to_nc(self.datum_cf_var_cdl)
+ with iris.FUTURE.context(datum_support=True):
+ cube = iris.load_cube(nc_path)
+ test_crs = cube.coord("projection_y_coordinate").coord_system
+ actual = str(test_crs.as_cartopy_crs().datum)
+ self.assertMultiLineEqual(expected, actual)
+
+ def test_no_load_datum_cf_var(self):
+ nc_path = tlc.cdl_to_nc(self.datum_cf_var_cdl)
+ with self.assertWarnsRegex(FutureWarning, "iris.FUTURE.datum_support"):
+ cube = iris.load_cube(nc_path)
+ test_crs = cube.coord("projection_y_coordinate").coord_system
+ actual = str(test_crs.as_cartopy_crs().datum)
+ self.assertMultiLineEqual(actual, "unknown")
+
+ def test_save_datum(self):
+ expected = "OSGB 1936"
+ saved_crs = iris.coord_systems.Mercator(
+ ellipsoid=iris.coord_systems.GeogCS.from_datum("OSGB36")
+ )
+
+ base_cube = stock.realistic_3d()
+ base_lat_coord = base_cube.coord("grid_latitude")
+ test_lat_coord = DimCoord(
+ base_lat_coord.points,
+ standard_name="projection_y_coordinate",
+ coord_system=saved_crs,
+ )
+ base_lon_coord = base_cube.coord("grid_longitude")
+ test_lon_coord = DimCoord(
+ base_lon_coord.points,
+ standard_name="projection_x_coordinate",
+ coord_system=saved_crs,
+ )
+ test_cube = Cube(
+ base_cube.data,
+ standard_name=base_cube.standard_name,
+ units=base_cube.units,
+ dim_coords_and_dims=(
+ (base_cube.coord("time"), 0),
+ (test_lat_coord, 1),
+ (test_lon_coord, 2),
+ ),
+ )
+
+ with self.temp_filename(suffix=".nc") as filename:
+ iris.save(test_cube, filename)
+ with iris.FUTURE.context(datum_support=True):
+ cube = iris.load_cube(filename)
+
+ test_crs = cube.coord("projection_y_coordinate").coord_system
+ actual = str(test_crs.as_cartopy_crs().datum)
+ self.assertMultiLineEqual(expected, actual)
+
+
+class TestLoadMinimalGeostationary(tests.IrisTest):
+ """
+ Check we can load data with a geostationary grid-mapping, even when the
+ 'false-easting' and 'false_northing' properties are missing.
+
+ """
+
+ _geostationary_problem_cdl = """
+netcdf geostationary_problem_case {
+dimensions:
+ y = 2 ;
+ x = 3 ;
+variables:
+ short radiance(y, x) ;
+ radiance:standard_name = "toa_outgoing_radiance_per_unit_wavelength" ;
+ radiance:units = "W m-2 sr-1 um-1" ;
+ radiance:coordinates = "y x" ;
+ radiance:grid_mapping = "imager_grid_mapping" ;
+ short y(y) ;
+ y:units = "rad" ;
+ y:axis = "Y" ;
+ y:long_name = "fixed grid projection y-coordinate" ;
+ y:standard_name = "projection_y_coordinate" ;
+ short x(x) ;
+ x:units = "rad" ;
+ x:axis = "X" ;
+ x:long_name = "fixed grid projection x-coordinate" ;
+ x:standard_name = "projection_x_coordinate" ;
+ int imager_grid_mapping ;
+ imager_grid_mapping:grid_mapping_name = "geostationary" ;
+ imager_grid_mapping:perspective_point_height = 35786023. ;
+ imager_grid_mapping:semi_major_axis = 6378137. ;
+ imager_grid_mapping:semi_minor_axis = 6356752.31414 ;
+ imager_grid_mapping:latitude_of_projection_origin = 0. ;
+ imager_grid_mapping:longitude_of_projection_origin = -75. ;
+ imager_grid_mapping:sweep_angle_axis = "x" ;
+
+data:
+
+ // coord values, just so these can be dim-coords
+ y = 0, 1 ;
+ x = 0, 1, 2 ;
+
+}
+"""
+
+ @classmethod
+ def setUpClass(cls):
+ # Create a temp directory for transient test files.
+ cls.temp_dir = tempfile.mkdtemp()
+ cls.path_test_cdl = path_join(cls.temp_dir, "geos_problem.cdl")
+ cls.path_test_nc = path_join(cls.temp_dir, "geos_problem.nc")
+ # Create reference CDL and netcdf files from the CDL text.
+ ncgen_from_cdl(
+ cdl_str=cls._geostationary_problem_cdl,
+ cdl_path=cls.path_test_cdl,
+ nc_path=cls.path_test_nc,
+ )
+
+ @classmethod
+ def tearDownClass(cls):
+ # Destroy the temp directory.
+ shutil.rmtree(cls.temp_dir)
+
+ def test_geostationary_no_false_offsets(self):
+ # Check we can load the test data and coordinate system properties are correct.
+ cube = iris.load_cube(self.path_test_nc)
+ # Check the coordinate system properties has the correct default properties.
+ cs = cube.coord_system()
+ self.assertIsInstance(cs, iris.coord_systems.Geostationary)
+ self.assertEqual(cs.false_easting, 0.0)
+ self.assertEqual(cs.false_northing, 0.0)
+
+
+if __name__ == "__main__":
+ tests.main()
diff --git a/lib/iris/tests/integration/netcdf/test_general.py b/lib/iris/tests/integration/netcdf/test_general.py
new file mode 100644
index 0000000000..63b977674d
--- /dev/null
+++ b/lib/iris/tests/integration/netcdf/test_general.py
@@ -0,0 +1,360 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""Integration tests for loading and saving netcdf files."""
+
+# Import iris.tests first so that some things can be initialised before
+# importing anything else.
+import iris.tests as tests # isort:skip
+
+from itertools import repeat
+import os.path
+import shutil
+import tempfile
+import warnings
+
+import numpy as np
+import numpy.ma as ma
+import pytest
+
+import iris
+import iris.coord_systems
+from iris.coords import CellMethod
+from iris.cube import Cube, CubeList
+import iris.exceptions
+from iris.fileformats.netcdf import Saver, UnknownCellMethodWarning
+from iris.tests.stock.netcdf import ncgen_from_cdl
+
+
+class TestLazySave(tests.IrisTest):
+ @tests.skip_data
+ def test_lazy_preserved_save(self):
+ fpath = tests.get_data_path(
+ ("NetCDF", "label_and_climate", "small_FC_167_mon_19601101.nc")
+ )
+ acube = iris.load_cube(fpath, "air_temperature")
+ self.assertTrue(acube.has_lazy_data())
+ # Also check a coord with lazy points + bounds.
+ self.assertTrue(acube.coord("forecast_period").has_lazy_points())
+ self.assertTrue(acube.coord("forecast_period").has_lazy_bounds())
+ with self.temp_filename(".nc") as nc_path:
+ with Saver(nc_path, "NETCDF4") as saver:
+ saver.write(acube)
+ # Check that cube data is not realised, also coord points + bounds.
+ self.assertTrue(acube.has_lazy_data())
+ self.assertTrue(acube.coord("forecast_period").has_lazy_points())
+ self.assertTrue(acube.coord("forecast_period").has_lazy_bounds())
+
+
+@tests.skip_data
+class TestCellMeasures(tests.IrisTest):
+ def setUp(self):
+ self.fname = tests.get_data_path(("NetCDF", "ORCA2", "votemper.nc"))
+
+ def test_load_raw(self):
+ (cube,) = iris.load_raw(self.fname)
+ self.assertEqual(len(cube.cell_measures()), 1)
+ self.assertEqual(cube.cell_measures()[0].measure, "area")
+
+ def test_load(self):
+ cube = iris.load_cube(self.fname)
+ self.assertEqual(len(cube.cell_measures()), 1)
+ self.assertEqual(cube.cell_measures()[0].measure, "area")
+
+ def test_merge_cell_measure_aware(self):
+ (cube1,) = iris.load_raw(self.fname)
+ (cube2,) = iris.load_raw(self.fname)
+ cube2._cell_measures_and_dims[0][0].var_name = "not_areat"
+ cubes = CubeList([cube1, cube2]).merge()
+ self.assertEqual(len(cubes), 2)
+
+ def test_concatenate_cell_measure_aware(self):
+ (cube1,) = iris.load_raw(self.fname)
+ cube1 = cube1[:, :, 0, 0]
+ cm_and_dims = cube1._cell_measures_and_dims
+ (cube2,) = iris.load_raw(self.fname)
+ cube2 = cube2[:, :, 0, 0]
+ cube2._cell_measures_and_dims[0][0].var_name = "not_areat"
+ cube2.coord("time").points = cube2.coord("time").points + 1
+ cubes = CubeList([cube1, cube2]).concatenate()
+ self.assertEqual(cubes[0]._cell_measures_and_dims, cm_and_dims)
+ self.assertEqual(len(cubes), 2)
+
+ def test_concatenate_cell_measure_match(self):
+ (cube1,) = iris.load_raw(self.fname)
+ cube1 = cube1[:, :, 0, 0]
+ cm_and_dims = cube1._cell_measures_and_dims
+ (cube2,) = iris.load_raw(self.fname)
+ cube2 = cube2[:, :, 0, 0]
+ cube2.coord("time").points = cube2.coord("time").points + 1
+ cubes = CubeList([cube1, cube2]).concatenate()
+ self.assertEqual(cubes[0]._cell_measures_and_dims, cm_and_dims)
+ self.assertEqual(len(cubes), 1)
+
+ def test_round_trip(self):
+ (cube,) = iris.load(self.fname)
+ with self.temp_filename(suffix=".nc") as filename:
+ iris.save(cube, filename, unlimited_dimensions=[])
+ (round_cube,) = iris.load_raw(filename)
+ self.assertEqual(len(round_cube.cell_measures()), 1)
+ self.assertEqual(round_cube.cell_measures()[0].measure, "area")
+
+ def test_print(self):
+ cube = iris.load_cube(self.fname)
+ printed = cube.__str__()
+ self.assertIn(
+ (
+ "Cell measures:\n"
+ " cell_area - - "
+ " x x"
+ ),
+ printed,
+ )
+
+
+class TestCellMethod_unknown(tests.IrisTest):
+ def test_unknown_method(self):
+ cube = Cube([1, 2], long_name="odd_phenomenon")
+ cube.add_cell_method(CellMethod(method="oddity", coords=("x",)))
+ temp_dirpath = tempfile.mkdtemp()
+ try:
+ temp_filepath = os.path.join(temp_dirpath, "tmp.nc")
+ iris.save(cube, temp_filepath)
+ with warnings.catch_warnings(record=True) as warning_records:
+ iris.load(temp_filepath)
+ # Filter to get the warning we are interested in.
+ warning_messages = [record.message for record in warning_records]
+ warning_messages = [
+ warn
+ for warn in warning_messages
+ if isinstance(warn, UnknownCellMethodWarning)
+ ]
+ self.assertEqual(len(warning_messages), 1)
+ message = warning_messages[0].args[0]
+ msg = (
+ "NetCDF variable 'odd_phenomenon' contains unknown cell "
+ "method 'oddity'"
+ )
+ self.assertIn(msg, message)
+ finally:
+ shutil.rmtree(temp_dirpath)
+
+
+def _get_scale_factor_add_offset(cube, datatype):
+ """Utility function used by netCDF data packing tests."""
+ if isinstance(datatype, dict):
+ dt = np.dtype(datatype["dtype"])
+ else:
+ dt = np.dtype(datatype)
+ cmax = cube.data.max()
+ cmin = cube.data.min()
+ n = dt.itemsize * 8
+ if ma.isMaskedArray(cube.data):
+ masked = True
+ else:
+ masked = False
+ if masked:
+ scale_factor = (cmax - cmin) / (2**n - 2)
+ else:
+ scale_factor = (cmax - cmin) / (2**n - 1)
+ if dt.kind == "u":
+ add_offset = cmin
+ elif dt.kind == "i":
+ if masked:
+ add_offset = (cmax + cmin) / 2
+ else:
+ add_offset = cmin + 2 ** (n - 1) * scale_factor
+ return (scale_factor, add_offset)
+
+
+@tests.skip_data
+class TestPackedData(tests.IrisTest):
+ def _single_test(self, datatype, CDLfilename, manual=False):
+ # Read PP input file.
+ file_in = tests.get_data_path(
+ (
+ "PP",
+ "cf_processing",
+ "000003000000.03.236.000128.1990.12.01.00.00.b.pp",
+ )
+ )
+ cube = iris.load_cube(file_in)
+ scale_factor, offset = _get_scale_factor_add_offset(cube, datatype)
+ if manual:
+ packspec = dict(
+ dtype=datatype, scale_factor=scale_factor, add_offset=offset
+ )
+ else:
+ packspec = datatype
+ # Write Cube to netCDF file.
+ with self.temp_filename(suffix=".nc") as file_out:
+ iris.save(cube, file_out, packing=packspec)
+ decimal = int(-np.log10(scale_factor))
+ packedcube = iris.load_cube(file_out)
+ # Check that packed cube is accurate to expected precision
+ self.assertArrayAlmostEqual(
+ cube.data, packedcube.data, decimal=decimal
+ )
+ # Check the netCDF file against CDL expected output.
+ self.assertCDL(
+ file_out,
+ (
+ "integration",
+ "netcdf",
+ "general",
+ "TestPackedData",
+ CDLfilename,
+ ),
+ )
+
+ def test_single_packed_signed(self):
+ """Test saving a single CF-netCDF file with packing."""
+ self._single_test("i2", "single_packed_signed.cdl")
+
+ def test_single_packed_unsigned(self):
+ """Test saving a single CF-netCDF file with packing into unsigned."""
+ self._single_test("u1", "single_packed_unsigned.cdl")
+
+ def test_single_packed_manual_scale(self):
+ """Test saving a single CF-netCDF file with packing with scale
+ factor and add_offset set manually."""
+ self._single_test("i2", "single_packed_manual.cdl", manual=True)
+
+ def _multi_test(self, CDLfilename, multi_dtype=False):
+ """Test saving multiple packed cubes with pack_dtype list."""
+ # Read PP input file.
+ file_in = tests.get_data_path(
+ ("PP", "cf_processing", "abcza_pa19591997_daily_29.b.pp")
+ )
+ cubes = iris.load(file_in)
+ # ensure cube order is the same:
+ cubes.sort(key=lambda cube: cube.cell_methods[0].method)
+ datatype = "i2"
+ scale_factor, offset = _get_scale_factor_add_offset(cubes[0], datatype)
+ if multi_dtype:
+ packdict = dict(
+ dtype=datatype, scale_factor=scale_factor, add_offset=offset
+ )
+ packspec = [packdict, None, "u2"]
+ dtypes = packspec
+ else:
+ packspec = datatype
+ dtypes = repeat(packspec)
+
+ # Write Cube to netCDF file.
+ with self.temp_filename(suffix=".nc") as file_out:
+ iris.save(cubes, file_out, packing=packspec)
+ # Check the netCDF file against CDL expected output.
+ self.assertCDL(
+ file_out,
+ (
+ "integration",
+ "netcdf",
+ "general",
+ "TestPackedData",
+ CDLfilename,
+ ),
+ )
+ packedcubes = iris.load(file_out)
+ packedcubes.sort(key=lambda cube: cube.cell_methods[0].method)
+ for cube, packedcube, dtype in zip(cubes, packedcubes, dtypes):
+ if dtype:
+ sf, ao = _get_scale_factor_add_offset(cube, dtype)
+ decimal = int(-np.log10(sf))
+ # Check that packed cube is accurate to expected precision
+ self.assertArrayAlmostEqual(
+ cube.data, packedcube.data, decimal=decimal
+ )
+ else:
+ self.assertArrayEqual(cube.data, packedcube.data)
+
+ def test_multi_packed_single_dtype(self):
+ """Test saving multiple packed cubes with the same pack_dtype."""
+ # Read PP input file.
+ self._multi_test("multi_packed_single_dtype.cdl")
+
+ def test_multi_packed_multi_dtype(self):
+ """Test saving multiple packed cubes with pack_dtype list."""
+ # Read PP input file.
+ self._multi_test("multi_packed_multi_dtype.cdl", multi_dtype=True)
+
+
+class TestScalarCube(tests.IrisTest):
+ def test_scalar_cube_save_load(self):
+ cube = iris.cube.Cube(1, long_name="scalar_cube")
+ with self.temp_filename(suffix=".nc") as fout:
+ iris.save(cube, fout)
+ scalar_cube = iris.load_cube(fout)
+ self.assertEqual(scalar_cube.name(), "scalar_cube")
+
+
+@tests.skip_data
+class TestConstrainedLoad(tests.IrisTest):
+ filename = tests.get_data_path(
+ ("NetCDF", "label_and_climate", "A1B-99999a-river-sep-2070-2099.nc")
+ )
+
+ def test_netcdf_with_NameConstraint(self):
+ constr = iris.NameConstraint(var_name="cdf_temp_dmax_tmean_abs")
+ cubes = iris.load(self.filename, constr)
+ self.assertEqual(len(cubes), 1)
+ self.assertEqual(cubes[0].var_name, "cdf_temp_dmax_tmean_abs")
+
+ def test_netcdf_with_no_constraint(self):
+ cubes = iris.load(self.filename)
+ self.assertEqual(len(cubes), 3)
+
+
+class TestSkippedCoord:
+ # If a coord/cell measure/etcetera cannot be added to the loaded Cube, a
+ # Warning is raised and the coord is skipped.
+ # This 'catching' is generic to all CannotAddErrors, but currently the only
+ # such problem that can exist in a NetCDF file is a mismatch of dimensions
+ # between phenomenon and coord.
+
+ cdl_core = """
+dimensions:
+ length_scale = 1 ;
+ lat = 3 ;
+variables:
+ float lat(lat) ;
+ lat:standard_name = "latitude" ;
+ lat:units = "degrees_north" ;
+ short lst_unc_sys(length_scale) ;
+ lst_unc_sys:long_name = "uncertainty from large-scale systematic
+ errors" ;
+ lst_unc_sys:units = "kelvin" ;
+ lst_unc_sys:coordinates = "lat" ;
+
+data:
+ lat = 0, 1, 2;
+ """
+
+ @pytest.fixture(autouse=True)
+ def create_nc_file(self, tmp_path):
+ file_name = "dim_mismatch"
+ cdl = f"netcdf {file_name}" + "{\n" + self.cdl_core + "\n}"
+ self.nc_path = (tmp_path / file_name).with_suffix(".nc")
+ ncgen_from_cdl(
+ cdl_str=cdl,
+ cdl_path=None,
+ nc_path=str(self.nc_path),
+ )
+ yield
+ self.nc_path.unlink()
+
+ def test_lat_not_loaded(self):
+ # iris#5068 includes discussion of possible retention of the skipped
+ # coords in the future.
+ with pytest.warns(
+ match="Missing data dimensions for multi-valued DimCoord"
+ ):
+ cube = iris.load_cube(self.nc_path)
+ with pytest.raises(iris.exceptions.CoordinateNotFoundError):
+ _ = cube.coord("lat")
+
+
+if __name__ == "__main__":
+ tests.main()
diff --git a/lib/iris/tests/integration/netcdf/test_self_referencing.py b/lib/iris/tests/integration/netcdf/test_self_referencing.py
new file mode 100644
index 0000000000..3395296e11
--- /dev/null
+++ b/lib/iris/tests/integration/netcdf/test_self_referencing.py
@@ -0,0 +1,126 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""Integration tests for iris#3367 - loading a self-referencing NetCDF file."""
+
+# Import iris.tests first so that some things can be initialised before
+# importing anything else.
+import iris.tests as tests # isort:skip
+
+import os
+import tempfile
+from unittest import mock
+
+import numpy as np
+
+import iris
+from iris.fileformats.netcdf import _thread_safe_nc
+
+
+@tests.skip_data
+class TestCMIP6VolcelloLoad(tests.IrisTest):
+ def setUp(self):
+ self.fname = tests.get_data_path(
+ (
+ "NetCDF",
+ "volcello",
+ "volcello_Ofx_CESM2_deforest-globe_r1i1p1f1_gn.nc",
+ )
+ )
+
+ def test_cmip6_volcello_load_issue_3367(self):
+ # Ensure that reading a file which references itself in
+ # `cell_measures` can be read. At the same time, ensure that we
+ # still receive a warning about other variables mentioned in
+ # `cell_measures` i.e. a warning should be raised about missing
+ # areacello.
+ areacello_str = "areacello"
+ volcello_str = "volcello"
+ expected_msg = (
+ "Missing CF-netCDF measure variable %r, "
+ "referenced by netCDF variable %r" % (areacello_str, volcello_str)
+ )
+
+ with mock.patch("warnings.warn") as warn:
+ # ensure file loads without failure
+ cube = iris.load_cube(self.fname)
+ warn.assert_has_calls([mock.call(expected_msg)])
+
+ # extra check to ensure correct variable was found
+ assert cube.standard_name == "ocean_volume"
+
+
+class TestSelfReferencingVarLoad(tests.IrisTest):
+ def setUp(self):
+ self.temp_dir_path = os.path.join(
+ tempfile.mkdtemp(), "issue_3367_volcello_test_file.nc"
+ )
+ dataset = _thread_safe_nc.DatasetWrapper(self.temp_dir_path, "w")
+
+ dataset.createDimension("lat", 4)
+ dataset.createDimension("lon", 5)
+ dataset.createDimension("lev", 3)
+
+ latitudes = dataset.createVariable("lat", np.float64, ("lat",))
+ longitudes = dataset.createVariable("lon", np.float64, ("lon",))
+ levels = dataset.createVariable("lev", np.float64, ("lev",))
+ volcello = dataset.createVariable(
+ "volcello", np.float32, ("lat", "lon", "lev")
+ )
+
+ latitudes.standard_name = "latitude"
+ latitudes.units = "degrees_north"
+ latitudes.axis = "Y"
+ latitudes[:] = np.linspace(-90, 90, 4)
+
+ longitudes.standard_name = "longitude"
+ longitudes.units = "degrees_east"
+ longitudes.axis = "X"
+ longitudes[:] = np.linspace(0, 360, 5)
+
+ levels.standard_name = "olevel"
+ levels.units = "centimeters"
+ levels.positive = "down"
+ levels.axis = "Z"
+ levels[:] = np.linspace(0, 10**5, 3)
+
+ volcello.id = "volcello"
+ volcello.out_name = "volcello"
+ volcello.standard_name = "ocean_volume"
+ volcello.units = "m3"
+ volcello.realm = "ocean"
+ volcello.frequency = "fx"
+ volcello.cell_measures = "area: areacello volume: volcello"
+ volcello = np.arange(4 * 5 * 3).reshape((4, 5, 3))
+
+ dataset.close()
+
+ def test_self_referencing_load_issue_3367(self):
+ # Ensure that reading a file which references itself in
+ # `cell_measures` can be read. At the same time, ensure that we
+ # still receive a warning about other variables mentioned in
+ # `cell_measures` i.e. a warning should be raised about missing
+ # areacello.
+ areacello_str = "areacello"
+ volcello_str = "volcello"
+ expected_msg = (
+ "Missing CF-netCDF measure variable %r, "
+ "referenced by netCDF variable %r" % (areacello_str, volcello_str)
+ )
+
+ with mock.patch("warnings.warn") as warn:
+ # ensure file loads without failure
+ cube = iris.load_cube(self.temp_dir_path)
+ warn.assert_called_with(expected_msg)
+
+ # extra check to ensure correct variable was found
+ assert cube.standard_name == "ocean_volume"
+
+ def tearDown(self):
+ os.remove(self.temp_dir_path)
+
+
+if __name__ == "__main__":
+ tests.main()
diff --git a/lib/iris/tests/integration/netcdf/test_thread_safety.py b/lib/iris/tests/integration/netcdf/test_thread_safety.py
new file mode 100644
index 0000000000..280e0f8418
--- /dev/null
+++ b/lib/iris/tests/integration/netcdf/test_thread_safety.py
@@ -0,0 +1,109 @@
+# Copyright Iris contributors
+#
+# This file is part of Iris and is released under the LGPL license.
+# See COPYING and COPYING.LESSER in the root of the repository for full
+# licensing details.
+"""
+Integration tests covering thread safety during loading/saving netcdf files.
+
+These tests are intended to catch non-thread-safe behaviour by producing CI
+'irregularities' that are noticed and investigated. They cannot reliably
+produce standard pytest failures, since the tools for 'correctly'
+testing non-thread-safe behaviour are not available at the Python layer.
+Thread safety problems can be either produce errors (like a normal test) OR
+segfaults (test doesn't complete, pytest-xdiff starts a new group worker, the
+end exit code is still non-0), and some problems do not occur in every test
+run.
+
+Token assertions are included after the line that is expected to reveal
+a thread safety problem, as this seems to be good testing practice.
+
+"""
+from pathlib import Path
+
+import dask
+from dask import array as da
+import numpy as np
+import pytest
+
+import iris
+from iris.cube import Cube, CubeList
+from iris.tests import get_data_path
+
+
+@pytest.fixture
+def tiny_chunks():
+ """Guarantee that Dask will use >1 thread by guaranteeing >1 chunk."""
+
+ def _check_tiny_loaded_chunks(cube: Cube):
+ assert cube.has_lazy_data()
+ cube_lazy_data = cube.core_data()
+ assert np.product(cube_lazy_data.chunksize) < cube_lazy_data.size
+
+ with dask.config.set({"array.chunk-size": "1KiB"}):
+ yield _check_tiny_loaded_chunks
+
+
+@pytest.fixture
+def save_common(tmp_path):
+ save_path = tmp_path / "tmp.nc"
+
+ def _func(cube: Cube):
+ assert not save_path.exists()
+ iris.save(cube, save_path)
+ assert save_path.exists()
+
+ yield _func
+
+
+@pytest.fixture
+def get_cubes_from_netcdf():
+ load_dir_path = Path(get_data_path(["NetCDF", "global", "xyt"]))
+ loaded = iris.load(load_dir_path.glob("*"), "tcco2")
+ smaller = CubeList([c[0] for c in loaded])
+ yield smaller
+
+
+def test_realise_data(tiny_chunks, get_cubes_from_netcdf):
+ cube = get_cubes_from_netcdf[0]
+ tiny_chunks(cube)
+ _ = cube.data # Any problems are expected here.
+ assert not cube.has_lazy_data()
+
+
+def test_realise_data_multisource(get_cubes_from_netcdf):
+ """Load from multiple sources to force Dask to use multiple threads."""
+ cubes = get_cubes_from_netcdf
+ final_cube = sum(cubes)
+ _ = final_cube.data # Any problems are expected here.
+ assert not final_cube.has_lazy_data()
+
+
+def test_save(tiny_chunks, save_common):
+ cube = Cube(da.ones(10000))
+ tiny_chunks(cube)
+ save_common(cube) # Any problems are expected here.
+
+
+def test_stream(tiny_chunks, get_cubes_from_netcdf, save_common):
+ cube = get_cubes_from_netcdf[0]
+ tiny_chunks(cube)
+ save_common(cube) # Any problems are expected here.
+
+
+def test_stream_multisource(get_cubes_from_netcdf, save_common):
+ """Load from multiple sources to force Dask to use multiple threads."""
+ cubes = get_cubes_from_netcdf
+ final_cube = sum(cubes)
+ save_common(final_cube) # Any problems are expected here.
+
+
+def test_comparison(get_cubes_from_netcdf):
+ """
+ Comparing multiple loaded files forces co-realisation.
+
+ See :func:`iris._lazy_data._co_realise_lazy_arrays` .
+ """
+ cubes = get_cubes_from_netcdf
+ _ = cubes[:-1] == cubes[1:] # Any problems are expected here.
+ assert all([c.has_lazy_data() for c in cubes])
diff --git a/lib/iris/tests/integration/plot/test_plot_2d_coords.py b/lib/iris/tests/integration/plot/test_plot_2d_coords.py
index b8fbc5e31a..1b95899803 100644
--- a/lib/iris/tests/integration/plot/test_plot_2d_coords.py
+++ b/lib/iris/tests/integration/plot/test_plot_2d_coords.py
@@ -38,10 +38,17 @@ def simple_cube_w_2d_coords():
class Test(tests.GraphicsTest):
def test_2d_coord_bounds_platecarree(self):
# To avoid a problem with Cartopy smearing the data where the
- # longitude wraps, we set the central_longitude
+ # longitude wraps, we set the central_longitude.
+ # SciTools/cartopy#1421
cube = simple_cube_w_2d_coords()[0, 0]
ax = plt.axes(projection=ccrs.PlateCarree(central_longitude=180))
qplt.pcolormesh(cube)
+
+ # Cartopy can't reliably set y-limits with curvilinear plotting.
+ # SciTools/cartopy#2121
+ y_lims = [m(cube.coord("latitude").points) for m in (np.min, np.max)]
+ ax.set_ylim(*y_lims)
+
ax.coastlines(resolution="110m", color="red")
self.check_graphic()
diff --git a/lib/iris/tests/integration/test_Datums.py b/lib/iris/tests/integration/test_Datums.py
index 77b7f28249..6953534f2d 100755
--- a/lib/iris/tests/integration/test_Datums.py
+++ b/lib/iris/tests/integration/test_Datums.py
@@ -23,7 +23,6 @@ def setUp(self):
self.start_crs = ccrs.OSGB(False)
def test_transform_points_datum(self):
-
# Iris version
wgs84 = GeogCS.from_datum("WGS84")
iris_cs = LambertConformal(
diff --git a/lib/iris/tests/integration/test_netcdf.py b/lib/iris/tests/integration/test_netcdf.py
deleted file mode 100644
index 851c539ade..0000000000
--- a/lib/iris/tests/integration/test_netcdf.py
+++ /dev/null
@@ -1,958 +0,0 @@
-# Copyright Iris contributors
-#
-# This file is part of Iris and is released under the LGPL license.
-# See COPYING and COPYING.LESSER in the root of the repository for full
-# licensing details.
-"""Integration tests for loading and saving netcdf files."""
-
-# Import iris.tests first so that some things can be initialised before
-# importing anything else.
-import iris.tests as tests # isort:skip
-
-from contextlib import contextmanager
-from itertools import repeat
-import os.path
-from os.path import join as path_join
-import shutil
-import tempfile
-from unittest import mock
-import warnings
-
-import netCDF4 as nc
-import numpy as np
-import numpy.ma as ma
-import pytest
-
-import iris
-import iris.coord_systems
-from iris.coords import CellMethod, DimCoord
-from iris.cube import Cube, CubeList
-import iris.exceptions
-from iris.fileformats.netcdf import (
- CF_CONVENTIONS_VERSION,
- Saver,
- UnknownCellMethodWarning,
-)
-import iris.tests.stock as stock
-from iris.tests.stock.netcdf import ncgen_from_cdl
-import iris.tests.unit.fileformats.netcdf.test_load_cubes as tlc
-
-
-@tests.skip_data
-class TestAtmosphereSigma(tests.IrisTest):
- def setUp(self):
- # Modify stock cube so it is suitable to have a atmosphere sigma
- # factory added to it.
- cube = stock.realistic_4d_no_derived()
- cube.coord("surface_altitude").rename("surface_air_pressure")
- cube.coord("surface_air_pressure").units = "Pa"
- cube.coord("sigma").units = "1"
- ptop_coord = iris.coords.AuxCoord(1000.0, var_name="ptop", units="Pa")
- cube.add_aux_coord(ptop_coord, ())
- cube.remove_coord("level_height")
- # Construct and add atmosphere sigma factory.
- factory = iris.aux_factory.AtmosphereSigmaFactory(
- cube.coord("ptop"),
- cube.coord("sigma"),
- cube.coord("surface_air_pressure"),
- )
- cube.add_aux_factory(factory)
- self.cube = cube
-
- def test_save(self):
- with self.temp_filename(suffix=".nc") as filename:
- iris.save(self.cube, filename)
- self.assertCDL(filename)
-
- def test_save_load_loop(self):
- # Ensure that the AtmosphereSigmaFactory is automatically loaded
- # when loading the file.
- with self.temp_filename(suffix=".nc") as filename:
- iris.save(self.cube, filename)
- cube = iris.load_cube(filename, "air_potential_temperature")
- assert cube.coords("air_pressure")
-
-
-@tests.skip_data
-class TestHybridPressure(tests.IrisTest):
- def setUp(self):
- # Modify stock cube so it is suitable to have a
- # hybrid pressure factory added to it.
- cube = stock.realistic_4d_no_derived()
- cube.coord("surface_altitude").rename("surface_air_pressure")
- cube.coord("surface_air_pressure").units = "Pa"
- cube.coord("level_height").rename("level_pressure")
- cube.coord("level_pressure").units = "Pa"
- # Construct and add hybrid pressure factory.
- factory = iris.aux_factory.HybridPressureFactory(
- cube.coord("level_pressure"),
- cube.coord("sigma"),
- cube.coord("surface_air_pressure"),
- )
- cube.add_aux_factory(factory)
- self.cube = cube
-
- def test_save(self):
- with self.temp_filename(suffix=".nc") as filename:
- iris.save(self.cube, filename)
- self.assertCDL(filename)
-
- def test_save_load_loop(self):
- # Tests an issue where the variable names in the formula
- # terms changed to the standard_names instead of the variable names
- # when loading a previously saved cube.
- with self.temp_filename(suffix=".nc") as filename, self.temp_filename(
- suffix=".nc"
- ) as other_filename:
- iris.save(self.cube, filename)
- cube = iris.load_cube(filename, "air_potential_temperature")
- iris.save(cube, other_filename)
- other_cube = iris.load_cube(
- other_filename, "air_potential_temperature"
- )
- self.assertEqual(cube, other_cube)
-
-
-@tests.skip_data
-class TestSaveMultipleAuxFactories(tests.IrisTest):
- def test_hybrid_height_and_pressure(self):
- cube = stock.realistic_4d()
- cube.add_aux_coord(
- iris.coords.DimCoord(
- 1200.0, long_name="level_pressure", units="hPa"
- )
- )
- cube.add_aux_coord(
- iris.coords.DimCoord(0.5, long_name="other sigma", units="1")
- )
- cube.add_aux_coord(
- iris.coords.DimCoord(
- 1000.0, long_name="surface_air_pressure", units="hPa"
- )
- )
- factory = iris.aux_factory.HybridPressureFactory(
- cube.coord("level_pressure"),
- cube.coord("other sigma"),
- cube.coord("surface_air_pressure"),
- )
- cube.add_aux_factory(factory)
- with self.temp_filename(suffix=".nc") as filename:
- iris.save(cube, filename)
- self.assertCDL(filename)
-
- def test_shared_primary(self):
- cube = stock.realistic_4d()
- factory = iris.aux_factory.HybridHeightFactory(
- cube.coord("level_height"),
- cube.coord("sigma"),
- cube.coord("surface_altitude"),
- )
- factory.rename("another altitude")
- cube.add_aux_factory(factory)
- with self.temp_filename(
- suffix=".nc"
- ) as filename, self.assertRaisesRegex(
- ValueError, "multiple aux factories"
- ):
- iris.save(cube, filename)
-
- def test_hybrid_height_cubes(self):
- hh1 = stock.simple_4d_with_hybrid_height()
- hh1.attributes["cube"] = "hh1"
- hh2 = stock.simple_4d_with_hybrid_height()
- hh2.attributes["cube"] = "hh2"
- sa = hh2.coord("surface_altitude")
- sa.points = sa.points * 10
- with self.temp_filename(".nc") as fname:
- iris.save([hh1, hh2], fname)
- cubes = iris.load(fname, "air_temperature")
- cubes = sorted(cubes, key=lambda cube: cube.attributes["cube"])
- self.assertCML(cubes)
-
- def test_hybrid_height_cubes_on_dimension_coordinate(self):
- hh1 = stock.hybrid_height()
- hh2 = stock.hybrid_height()
- sa = hh2.coord("surface_altitude")
- sa.points = sa.points * 10
- emsg = "Unable to create dimensonless vertical coordinate."
- with self.temp_filename(".nc") as fname, self.assertRaisesRegex(
- ValueError, emsg
- ):
- iris.save([hh1, hh2], fname)
-
-
-class TestUmVersionAttribute(tests.IrisTest):
- def test_single_saves_as_global(self):
- cube = Cube(
- [1.0],
- standard_name="air_temperature",
- units="K",
- attributes={"um_version": "4.3"},
- )
- with self.temp_filename(".nc") as nc_path:
- iris.save(cube, nc_path)
- self.assertCDL(nc_path)
-
- def test_multiple_same_saves_as_global(self):
- cube_a = Cube(
- [1.0],
- standard_name="air_temperature",
- units="K",
- attributes={"um_version": "4.3"},
- )
- cube_b = Cube(
- [1.0],
- standard_name="air_pressure",
- units="hPa",
- attributes={"um_version": "4.3"},
- )
- with self.temp_filename(".nc") as nc_path:
- iris.save(CubeList([cube_a, cube_b]), nc_path)
- self.assertCDL(nc_path)
-
- def test_multiple_different_saves_on_variables(self):
- cube_a = Cube(
- [1.0],
- standard_name="air_temperature",
- units="K",
- attributes={"um_version": "4.3"},
- )
- cube_b = Cube(
- [1.0],
- standard_name="air_pressure",
- units="hPa",
- attributes={"um_version": "4.4"},
- )
- with self.temp_filename(".nc") as nc_path:
- iris.save(CubeList([cube_a, cube_b]), nc_path)
- self.assertCDL(nc_path)
-
-
-@contextmanager
-def _patch_site_configuration():
- def cf_patch_conventions(conventions):
- return ", ".join([conventions, "convention1, convention2"])
-
- def update(config):
- config["cf_profile"] = mock.Mock(name="cf_profile")
- config["cf_patch"] = mock.Mock(name="cf_patch")
- config["cf_patch_conventions"] = cf_patch_conventions
-
- orig_site_config = iris.site_configuration.copy()
- update(iris.site_configuration)
- yield
- iris.site_configuration = orig_site_config
-
-
-class TestConventionsAttributes(tests.IrisTest):
- def test_patching_conventions_attribute(self):
- # Ensure that user defined conventions are wiped and those which are
- # saved patched through site_config can be loaded without an exception
- # being raised.
- cube = Cube(
- [1.0],
- standard_name="air_temperature",
- units="K",
- attributes={"Conventions": "some user defined conventions"},
- )
-
- # Patch the site configuration dictionary.
- with _patch_site_configuration(), self.temp_filename(".nc") as nc_path:
- iris.save(cube, nc_path)
- res = iris.load_cube(nc_path)
-
- self.assertEqual(
- res.attributes["Conventions"],
- "{}, {}, {}".format(
- CF_CONVENTIONS_VERSION, "convention1", "convention2"
- ),
- )
-
-
-class TestLazySave(tests.IrisTest):
- @tests.skip_data
- def test_lazy_preserved_save(self):
- fpath = tests.get_data_path(
- ("NetCDF", "label_and_climate", "small_FC_167_mon_19601101.nc")
- )
- acube = iris.load_cube(fpath, "air_temperature")
- self.assertTrue(acube.has_lazy_data())
- # Also check a coord with lazy points + bounds.
- self.assertTrue(acube.coord("forecast_period").has_lazy_points())
- self.assertTrue(acube.coord("forecast_period").has_lazy_bounds())
- with self.temp_filename(".nc") as nc_path:
- with Saver(nc_path, "NETCDF4") as saver:
- saver.write(acube)
- # Check that cube data is not realised, also coord points + bounds.
- self.assertTrue(acube.has_lazy_data())
- self.assertTrue(acube.coord("forecast_period").has_lazy_points())
- self.assertTrue(acube.coord("forecast_period").has_lazy_bounds())
-
-
-@tests.skip_data
-class TestCellMeasures(tests.IrisTest):
- def setUp(self):
- self.fname = tests.get_data_path(("NetCDF", "ORCA2", "votemper.nc"))
-
- def test_load_raw(self):
- (cube,) = iris.load_raw(self.fname)
- self.assertEqual(len(cube.cell_measures()), 1)
- self.assertEqual(cube.cell_measures()[0].measure, "area")
-
- def test_load(self):
- cube = iris.load_cube(self.fname)
- self.assertEqual(len(cube.cell_measures()), 1)
- self.assertEqual(cube.cell_measures()[0].measure, "area")
-
- def test_merge_cell_measure_aware(self):
- (cube1,) = iris.load_raw(self.fname)
- (cube2,) = iris.load_raw(self.fname)
- cube2._cell_measures_and_dims[0][0].var_name = "not_areat"
- cubes = CubeList([cube1, cube2]).merge()
- self.assertEqual(len(cubes), 2)
-
- def test_concatenate_cell_measure_aware(self):
- (cube1,) = iris.load_raw(self.fname)
- cube1 = cube1[:, :, 0, 0]
- cm_and_dims = cube1._cell_measures_and_dims
- (cube2,) = iris.load_raw(self.fname)
- cube2 = cube2[:, :, 0, 0]
- cube2._cell_measures_and_dims[0][0].var_name = "not_areat"
- cube2.coord("time").points = cube2.coord("time").points + 1
- cubes = CubeList([cube1, cube2]).concatenate()
- self.assertEqual(cubes[0]._cell_measures_and_dims, cm_and_dims)
- self.assertEqual(len(cubes), 2)
-
- def test_concatenate_cell_measure_match(self):
- (cube1,) = iris.load_raw(self.fname)
- cube1 = cube1[:, :, 0, 0]
- cm_and_dims = cube1._cell_measures_and_dims
- (cube2,) = iris.load_raw(self.fname)
- cube2 = cube2[:, :, 0, 0]
- cube2.coord("time").points = cube2.coord("time").points + 1
- cubes = CubeList([cube1, cube2]).concatenate()
- self.assertEqual(cubes[0]._cell_measures_and_dims, cm_and_dims)
- self.assertEqual(len(cubes), 1)
-
- def test_round_trip(self):
- (cube,) = iris.load(self.fname)
- with self.temp_filename(suffix=".nc") as filename:
- iris.save(cube, filename, unlimited_dimensions=[])
- (round_cube,) = iris.load_raw(filename)
- self.assertEqual(len(round_cube.cell_measures()), 1)
- self.assertEqual(round_cube.cell_measures()[0].measure, "area")
-
- def test_print(self):
- cube = iris.load_cube(self.fname)
- printed = cube.__str__()
- self.assertIn(
- (
- "Cell measures:\n"
- " cell_area - - "
- " x x"
- ),
- printed,
- )
-
-
-@tests.skip_data
-class TestCMIP6VolcelloLoad(tests.IrisTest):
- def setUp(self):
- self.fname = tests.get_data_path(
- (
- "NetCDF",
- "volcello",
- "volcello_Ofx_CESM2_deforest-globe_r1i1p1f1_gn.nc",
- )
- )
-
- def test_cmip6_volcello_load_issue_3367(self):
- # Ensure that reading a file which references itself in
- # `cell_measures` can be read. At the same time, ensure that we
- # still receive a warning about other variables mentioned in
- # `cell_measures` i.e. a warning should be raised about missing
- # areacello.
- areacello_str = "areacello"
- volcello_str = "volcello"
- expected_msg = (
- "Missing CF-netCDF measure variable %r, "
- "referenced by netCDF variable %r" % (areacello_str, volcello_str)
- )
-
- with mock.patch("warnings.warn") as warn:
- # ensure file loads without failure
- cube = iris.load_cube(self.fname)
- warn.assert_has_calls([mock.call(expected_msg)])
-
- # extra check to ensure correct variable was found
- assert cube.standard_name == "ocean_volume"
-
-
-class TestSelfReferencingVarLoad(tests.IrisTest):
- def setUp(self):
- self.temp_dir_path = os.path.join(
- tempfile.mkdtemp(), "issue_3367_volcello_test_file.nc"
- )
- dataset = nc.Dataset(self.temp_dir_path, "w")
-
- dataset.createDimension("lat", 4)
- dataset.createDimension("lon", 5)
- dataset.createDimension("lev", 3)
-
- latitudes = dataset.createVariable("lat", np.float64, ("lat",))
- longitudes = dataset.createVariable("lon", np.float64, ("lon",))
- levels = dataset.createVariable("lev", np.float64, ("lev",))
- volcello = dataset.createVariable(
- "volcello", np.float32, ("lat", "lon", "lev")
- )
-
- latitudes.standard_name = "latitude"
- latitudes.units = "degrees_north"
- latitudes.axis = "Y"
- latitudes[:] = np.linspace(-90, 90, 4)
-
- longitudes.standard_name = "longitude"
- longitudes.units = "degrees_east"
- longitudes.axis = "X"
- longitudes[:] = np.linspace(0, 360, 5)
-
- levels.standard_name = "olevel"
- levels.units = "centimeters"
- levels.positive = "down"
- levels.axis = "Z"
- levels[:] = np.linspace(0, 10**5, 3)
-
- volcello.id = "volcello"
- volcello.out_name = "volcello"
- volcello.standard_name = "ocean_volume"
- volcello.units = "m3"
- volcello.realm = "ocean"
- volcello.frequency = "fx"
- volcello.cell_measures = "area: areacello volume: volcello"
- volcello = np.arange(4 * 5 * 3).reshape((4, 5, 3))
-
- dataset.close()
-
- def test_self_referencing_load_issue_3367(self):
- # Ensure that reading a file which references itself in
- # `cell_measures` can be read. At the same time, ensure that we
- # still receive a warning about other variables mentioned in
- # `cell_measures` i.e. a warning should be raised about missing
- # areacello.
- areacello_str = "areacello"
- volcello_str = "volcello"
- expected_msg = (
- "Missing CF-netCDF measure variable %r, "
- "referenced by netCDF variable %r" % (areacello_str, volcello_str)
- )
-
- with mock.patch("warnings.warn") as warn:
- # ensure file loads without failure
- cube = iris.load_cube(self.temp_dir_path)
- warn.assert_called_with(expected_msg)
-
- # extra check to ensure correct variable was found
- assert cube.standard_name == "ocean_volume"
-
- def tearDown(self):
- os.remove(self.temp_dir_path)
-
-
-class TestCellMethod_unknown(tests.IrisTest):
- def test_unknown_method(self):
- cube = Cube([1, 2], long_name="odd_phenomenon")
- cube.add_cell_method(CellMethod(method="oddity", coords=("x",)))
- temp_dirpath = tempfile.mkdtemp()
- try:
- temp_filepath = os.path.join(temp_dirpath, "tmp.nc")
- iris.save(cube, temp_filepath)
- with warnings.catch_warnings(record=True) as warning_records:
- iris.load(temp_filepath)
- # Filter to get the warning we are interested in.
- warning_messages = [record.message for record in warning_records]
- warning_messages = [
- warn
- for warn in warning_messages
- if isinstance(warn, UnknownCellMethodWarning)
- ]
- self.assertEqual(len(warning_messages), 1)
- message = warning_messages[0].args[0]
- msg = (
- "NetCDF variable 'odd_phenomenon' contains unknown cell "
- "method 'oddity'"
- )
- self.assertIn(msg, message)
- finally:
- shutil.rmtree(temp_dirpath)
-
-
-@tests.skip_data
-class TestCoordSystem(tests.IrisTest):
- def setUp(self):
- tlc.setUpModule()
-
- def tearDown(self):
- tlc.tearDownModule()
-
- def test_load_laea_grid(self):
- cube = iris.load_cube(
- tests.get_data_path(
- ("NetCDF", "lambert_azimuthal_equal_area", "euro_air_temp.nc")
- )
- )
- self.assertCML(cube, ("netcdf", "netcdf_laea.cml"))
-
- datum_cf_var_cdl = """
- netcdf output {
- dimensions:
- y = 4 ;
- x = 3 ;
- variables:
- float data(y, x) ;
- data :standard_name = "toa_brightness_temperature" ;
- data :units = "K" ;
- data :grid_mapping = "mercator" ;
- int mercator ;
- mercator:grid_mapping_name = "mercator" ;
- mercator:longitude_of_prime_meridian = 0. ;
- mercator:earth_radius = 6378169. ;
- mercator:horizontal_datum_name = "OSGB36" ;
- float y(y) ;
- y:axis = "Y" ;
- y:units = "m" ;
- y:standard_name = "projection_y_coordinate" ;
- float x(x) ;
- x:axis = "X" ;
- x:units = "m" ;
- x:standard_name = "projection_x_coordinate" ;
-
- // global attributes:
- :Conventions = "CF-1.7" ;
- :standard_name_vocabulary = "CF Standard Name Table v27" ;
-
- data:
-
- data =
- 0, 1, 2,
- 3, 4, 5,
- 6, 7, 8,
- 9, 10, 11 ;
-
- mercator = _ ;
-
- y = 1, 2, 3, 5 ;
-
- x = -6, -4, -2 ;
-
- }
- """
-
- datum_wkt_cdl = """
-netcdf output5 {
-dimensions:
- y = 4 ;
- x = 3 ;
-variables:
- float data(y, x) ;
- data :standard_name = "toa_brightness_temperature" ;
- data :units = "K" ;
- data :grid_mapping = "mercator" ;
- int mercator ;
- mercator:grid_mapping_name = "mercator" ;
- mercator:longitude_of_prime_meridian = 0. ;
- mercator:earth_radius = 6378169. ;
- mercator:longitude_of_projection_origin = 0. ;
- mercator:false_easting = 0. ;
- mercator:false_northing = 0. ;
- mercator:scale_factor_at_projection_origin = 1. ;
- mercator:crs_wkt = "PROJCRS[\\"unknown\\",BASEGEOGCRS[\\"unknown\\",DATUM[\\"OSGB36\\",ELLIPSOID[\\"unknown\\",6378169,0,LENGTHUNIT[\\"metre\\",1,ID[\\"EPSG\\",9001]]]],PRIMEM[\\"Greenwich\\",0,ANGLEUNIT[\\"degree\\",0.0174532925199433],ID[\\"EPSG\\",8901]]],CONVERSION[\\"unknown\\",METHOD[\\"Mercator (variant B)\\",ID[\\"EPSG\\",9805]],PARAMETER[\\"Latitude of 1st standard parallel\\",0,ANGLEUNIT[\\"degree\\",0.0174532925199433],ID[\\"EPSG\\",8823]],PARAMETER[\\"Longitude of natural origin\\",0,ANGLEUNIT[\\"degree\\",0.0174532925199433],ID[\\"EPSG\\",8802]],PARAMETER[\\"False easting\\",0,LENGTHUNIT[\\"metre\\",1],ID[\\"EPSG\\",8806]],PARAMETER[\\"False northing\\",0,LENGTHUNIT[\\"metre\\",1],ID[\\"EPSG\\",8807]]],CS[Cartesian,2],AXIS[\\"(E)\\",east,ORDER[1],LENGTHUNIT[\\"metre\\",1,ID[\\"EPSG\\",9001]]],AXIS[\\"(N)\\",north,ORDER[2],LENGTHUNIT[\\"metre\\",1,ID[\\"EPSG\\",9001]]]]" ;
- float y(y) ;
- y:axis = "Y" ;
- y:units = "m" ;
- y:standard_name = "projection_y_coordinate" ;
- float x(x) ;
- x:axis = "X" ;
- x:units = "m" ;
- x:standard_name = "projection_x_coordinate" ;
-
-// global attributes:
- :standard_name_vocabulary = "CF Standard Name Table v27" ;
- :Conventions = "CF-1.7" ;
-data:
-
- data =
- 0, 1, 2,
- 3, 4, 5,
- 6, 7, 8,
- 9, 10, 11 ;
-
- mercator = _ ;
-
- y = 1, 2, 3, 5 ;
-
- x = -6, -4, -2 ;
-}
- """
-
- def test_load_datum_wkt(self):
- expected = "OSGB 1936"
- nc_path = tlc.cdl_to_nc(self.datum_wkt_cdl)
- with iris.FUTURE.context(datum_support=True):
- cube = iris.load_cube(nc_path)
- test_crs = cube.coord("projection_y_coordinate").coord_system
- actual = str(test_crs.as_cartopy_crs().datum)
- self.assertMultiLineEqual(expected, actual)
-
- def test_no_load_datum_wkt(self):
- nc_path = tlc.cdl_to_nc(self.datum_wkt_cdl)
- with self.assertWarnsRegex(FutureWarning, "iris.FUTURE.datum_support"):
- cube = iris.load_cube(nc_path)
- test_crs = cube.coord("projection_y_coordinate").coord_system
- actual = str(test_crs.as_cartopy_crs().datum)
- self.assertMultiLineEqual(actual, "unknown")
-
- def test_load_datum_cf_var(self):
- expected = "OSGB 1936"
- nc_path = tlc.cdl_to_nc(self.datum_cf_var_cdl)
- with iris.FUTURE.context(datum_support=True):
- cube = iris.load_cube(nc_path)
- test_crs = cube.coord("projection_y_coordinate").coord_system
- actual = str(test_crs.as_cartopy_crs().datum)
- self.assertMultiLineEqual(expected, actual)
-
- def test_no_load_datum_cf_var(self):
- nc_path = tlc.cdl_to_nc(self.datum_cf_var_cdl)
- with self.assertWarnsRegex(FutureWarning, "iris.FUTURE.datum_support"):
- cube = iris.load_cube(nc_path)
- test_crs = cube.coord("projection_y_coordinate").coord_system
- actual = str(test_crs.as_cartopy_crs().datum)
- self.assertMultiLineEqual(actual, "unknown")
-
- def test_save_datum(self):
- expected = "OSGB 1936"
- saved_crs = iris.coord_systems.Mercator(
- ellipsoid=iris.coord_systems.GeogCS.from_datum("OSGB36")
- )
-
- base_cube = stock.realistic_3d()
- base_lat_coord = base_cube.coord("grid_latitude")
- test_lat_coord = DimCoord(
- base_lat_coord.points,
- standard_name="projection_y_coordinate",
- coord_system=saved_crs,
- )
- base_lon_coord = base_cube.coord("grid_longitude")
- test_lon_coord = DimCoord(
- base_lon_coord.points,
- standard_name="projection_x_coordinate",
- coord_system=saved_crs,
- )
- test_cube = Cube(
- base_cube.data,
- standard_name=base_cube.standard_name,
- units=base_cube.units,
- dim_coords_and_dims=(
- (base_cube.coord("time"), 0),
- (test_lat_coord, 1),
- (test_lon_coord, 2),
- ),
- )
-
- with self.temp_filename(suffix=".nc") as filename:
- iris.save(test_cube, filename)
- with iris.FUTURE.context(datum_support=True):
- cube = iris.load_cube(filename)
-
- test_crs = cube.coord("projection_y_coordinate").coord_system
- actual = str(test_crs.as_cartopy_crs().datum)
- self.assertMultiLineEqual(expected, actual)
-
-
-def _get_scale_factor_add_offset(cube, datatype):
- """Utility function used by netCDF data packing tests."""
- if isinstance(datatype, dict):
- dt = np.dtype(datatype["dtype"])
- else:
- dt = np.dtype(datatype)
- cmax = cube.data.max()
- cmin = cube.data.min()
- n = dt.itemsize * 8
- if ma.isMaskedArray(cube.data):
- masked = True
- else:
- masked = False
- if masked:
- scale_factor = (cmax - cmin) / (2**n - 2)
- else:
- scale_factor = (cmax - cmin) / (2**n - 1)
- if dt.kind == "u":
- add_offset = cmin
- elif dt.kind == "i":
- if masked:
- add_offset = (cmax + cmin) / 2
- else:
- add_offset = cmin + 2 ** (n - 1) * scale_factor
- return (scale_factor, add_offset)
-
-
-@tests.skip_data
-class TestPackedData(tests.IrisTest):
- def _single_test(self, datatype, CDLfilename, manual=False):
- # Read PP input file.
- file_in = tests.get_data_path(
- (
- "PP",
- "cf_processing",
- "000003000000.03.236.000128.1990.12.01.00.00.b.pp",
- )
- )
- cube = iris.load_cube(file_in)
- scale_factor, offset = _get_scale_factor_add_offset(cube, datatype)
- if manual:
- packspec = dict(
- dtype=datatype, scale_factor=scale_factor, add_offset=offset
- )
- else:
- packspec = datatype
- # Write Cube to netCDF file.
- with self.temp_filename(suffix=".nc") as file_out:
- iris.save(cube, file_out, packing=packspec)
- decimal = int(-np.log10(scale_factor))
- packedcube = iris.load_cube(file_out)
- # Check that packed cube is accurate to expected precision
- self.assertArrayAlmostEqual(
- cube.data, packedcube.data, decimal=decimal
- )
- # Check the netCDF file against CDL expected output.
- self.assertCDL(
- file_out,
- ("integration", "netcdf", "TestPackedData", CDLfilename),
- )
-
- def test_single_packed_signed(self):
- """Test saving a single CF-netCDF file with packing."""
- self._single_test("i2", "single_packed_signed.cdl")
-
- def test_single_packed_unsigned(self):
- """Test saving a single CF-netCDF file with packing into unsigned."""
- self._single_test("u1", "single_packed_unsigned.cdl")
-
- def test_single_packed_manual_scale(self):
- """Test saving a single CF-netCDF file with packing with scale
- factor and add_offset set manually."""
- self._single_test("i2", "single_packed_manual.cdl", manual=True)
-
- def _multi_test(self, CDLfilename, multi_dtype=False):
- """Test saving multiple packed cubes with pack_dtype list."""
- # Read PP input file.
- file_in = tests.get_data_path(
- ("PP", "cf_processing", "abcza_pa19591997_daily_29.b.pp")
- )
- cubes = iris.load(file_in)
- # ensure cube order is the same:
- cubes.sort(key=lambda cube: cube.cell_methods[0].method)
- datatype = "i2"
- scale_factor, offset = _get_scale_factor_add_offset(cubes[0], datatype)
- if multi_dtype:
- packdict = dict(
- dtype=datatype, scale_factor=scale_factor, add_offset=offset
- )
- packspec = [packdict, None, "u2"]
- dtypes = packspec
- else:
- packspec = datatype
- dtypes = repeat(packspec)
-
- # Write Cube to netCDF file.
- with self.temp_filename(suffix=".nc") as file_out:
- iris.save(cubes, file_out, packing=packspec)
- # Check the netCDF file against CDL expected output.
- self.assertCDL(
- file_out,
- ("integration", "netcdf", "TestPackedData", CDLfilename),
- )
- packedcubes = iris.load(file_out)
- packedcubes.sort(key=lambda cube: cube.cell_methods[0].method)
- for cube, packedcube, dtype in zip(cubes, packedcubes, dtypes):
- if dtype:
- sf, ao = _get_scale_factor_add_offset(cube, dtype)
- decimal = int(-np.log10(sf))
- # Check that packed cube is accurate to expected precision
- self.assertArrayAlmostEqual(
- cube.data, packedcube.data, decimal=decimal
- )
- else:
- self.assertArrayEqual(cube.data, packedcube.data)
-
- def test_multi_packed_single_dtype(self):
- """Test saving multiple packed cubes with the same pack_dtype."""
- # Read PP input file.
- self._multi_test("multi_packed_single_dtype.cdl")
-
- def test_multi_packed_multi_dtype(self):
- """Test saving multiple packed cubes with pack_dtype list."""
- # Read PP input file.
- self._multi_test("multi_packed_multi_dtype.cdl", multi_dtype=True)
-
-
-class TestScalarCube(tests.IrisTest):
- def test_scalar_cube_save_load(self):
- cube = iris.cube.Cube(1, long_name="scalar_cube")
- with self.temp_filename(suffix=".nc") as fout:
- iris.save(cube, fout)
- scalar_cube = iris.load_cube(fout)
- self.assertEqual(scalar_cube.name(), "scalar_cube")
-
-
-class TestStandardName(tests.IrisTest):
- def test_standard_name_roundtrip(self):
- standard_name = "air_temperature detection_minimum"
- cube = iris.cube.Cube(1, standard_name=standard_name)
- with self.temp_filename(suffix=".nc") as fout:
- iris.save(cube, fout)
- detection_limit_cube = iris.load_cube(fout)
- self.assertEqual(detection_limit_cube.standard_name, standard_name)
-
-
-class TestLoadMinimalGeostationary(tests.IrisTest):
- """
- Check we can load data with a geostationary grid-mapping, even when the
- 'false-easting' and 'false_northing' properties are missing.
-
- """
-
- _geostationary_problem_cdl = """
-netcdf geostationary_problem_case {
-dimensions:
- y = 2 ;
- x = 3 ;
-variables:
- short radiance(y, x) ;
- radiance:standard_name = "toa_outgoing_radiance_per_unit_wavelength" ;
- radiance:units = "W m-2 sr-1 um-1" ;
- radiance:coordinates = "y x" ;
- radiance:grid_mapping = "imager_grid_mapping" ;
- short y(y) ;
- y:units = "rad" ;
- y:axis = "Y" ;
- y:long_name = "fixed grid projection y-coordinate" ;
- y:standard_name = "projection_y_coordinate" ;
- short x(x) ;
- x:units = "rad" ;
- x:axis = "X" ;
- x:long_name = "fixed grid projection x-coordinate" ;
- x:standard_name = "projection_x_coordinate" ;
- int imager_grid_mapping ;
- imager_grid_mapping:grid_mapping_name = "geostationary" ;
- imager_grid_mapping:perspective_point_height = 35786023. ;
- imager_grid_mapping:semi_major_axis = 6378137. ;
- imager_grid_mapping:semi_minor_axis = 6356752.31414 ;
- imager_grid_mapping:latitude_of_projection_origin = 0. ;
- imager_grid_mapping:longitude_of_projection_origin = -75. ;
- imager_grid_mapping:sweep_angle_axis = "x" ;
-
-data:
-
- // coord values, just so these can be dim-coords
- y = 0, 1 ;
- x = 0, 1, 2 ;
-
-}
-"""
-
- @classmethod
- def setUpClass(cls):
- # Create a temp directory for transient test files.
- cls.temp_dir = tempfile.mkdtemp()
- cls.path_test_cdl = path_join(cls.temp_dir, "geos_problem.cdl")
- cls.path_test_nc = path_join(cls.temp_dir, "geos_problem.nc")
- # Create reference CDL and netcdf files from the CDL text.
- ncgen_from_cdl(
- cdl_str=cls._geostationary_problem_cdl,
- cdl_path=cls.path_test_cdl,
- nc_path=cls.path_test_nc,
- )
-
- @classmethod
- def tearDownClass(cls):
- # Destroy the temp directory.
- shutil.rmtree(cls.temp_dir)
-
- def test_geostationary_no_false_offsets(self):
- # Check we can load the test data and coordinate system properties are correct.
- cube = iris.load_cube(self.path_test_nc)
- # Check the coordinate system properties has the correct default properties.
- cs = cube.coord_system()
- self.assertIsInstance(cs, iris.coord_systems.Geostationary)
- self.assertEqual(cs.false_easting, 0.0)
- self.assertEqual(cs.false_northing, 0.0)
-
-
-@tests.skip_data
-class TestConstrainedLoad(tests.IrisTest):
- filename = tests.get_data_path(
- ("NetCDF", "label_and_climate", "A1B-99999a-river-sep-2070-2099.nc")
- )
-
- def test_netcdf_with_NameConstraint(self):
- constr = iris.NameConstraint(var_name="cdf_temp_dmax_tmean_abs")
- cubes = iris.load(self.filename, constr)
- self.assertEqual(len(cubes), 1)
- self.assertEqual(cubes[0].var_name, "cdf_temp_dmax_tmean_abs")
-
- def test_netcdf_with_no_constraint(self):
- cubes = iris.load(self.filename)
- self.assertEqual(len(cubes), 3)
-
-
-class TestSkippedCoord:
- # If a coord/cell measure/etcetera cannot be added to the loaded Cube, a
- # Warning is raised and the coord is skipped.
- # This 'catching' is generic to all CannotAddErrors, but currently the only
- # such problem that can exist in a NetCDF file is a mismatch of dimensions
- # between phenomenon and coord.
-
- cdl_core = """
-dimensions:
- length_scale = 1 ;
- lat = 3 ;
-variables:
- float lat(lat) ;
- lat:standard_name = "latitude" ;
- lat:units = "degrees_north" ;
- short lst_unc_sys(length_scale) ;
- lst_unc_sys:long_name = "uncertainty from large-scale systematic
- errors" ;
- lst_unc_sys:units = "kelvin" ;
- lst_unc_sys:coordinates = "lat" ;
-
-data:
- lat = 0, 1, 2;
- """
-
- @pytest.fixture(autouse=True)
- def create_nc_file(self, tmp_path):
- file_name = "dim_mismatch"
- cdl = f"netcdf {file_name}" + "{\n" + self.cdl_core + "\n}"
- self.nc_path = (tmp_path / file_name).with_suffix(".nc")
- ncgen_from_cdl(
- cdl_str=cdl,
- cdl_path=None,
- nc_path=str(self.nc_path),
- )
- yield
- self.nc_path.unlink()
-
- def test_lat_not_loaded(self):
- # iris#5068 includes discussion of possible retention of the skipped
- # coords in the future.
- with pytest.warns(
- match="Missing data dimensions for multi-valued DimCoord"
- ):
- cube = iris.load_cube(self.nc_path)
- with pytest.raises(iris.exceptions.CoordinateNotFoundError):
- _ = cube.coord("lat")
-
-
-if __name__ == "__main__":
- tests.main()
diff --git a/lib/iris/tests/results/integration/netcdf/TestUmVersionAttribute/multiple_different_saves_on_variables.cdl b/lib/iris/tests/results/integration/netcdf/attributes/TestUmVersionAttribute/multiple_different_saves_on_variables.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestUmVersionAttribute/multiple_different_saves_on_variables.cdl
rename to lib/iris/tests/results/integration/netcdf/attributes/TestUmVersionAttribute/multiple_different_saves_on_variables.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestUmVersionAttribute/multiple_same_saves_as_global.cdl b/lib/iris/tests/results/integration/netcdf/attributes/TestUmVersionAttribute/multiple_same_saves_as_global.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestUmVersionAttribute/multiple_same_saves_as_global.cdl
rename to lib/iris/tests/results/integration/netcdf/attributes/TestUmVersionAttribute/multiple_same_saves_as_global.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestUmVersionAttribute/single_saves_as_global.cdl b/lib/iris/tests/results/integration/netcdf/attributes/TestUmVersionAttribute/single_saves_as_global.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestUmVersionAttribute/single_saves_as_global.cdl
rename to lib/iris/tests/results/integration/netcdf/attributes/TestUmVersionAttribute/single_saves_as_global.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestAtmosphereSigma/save.cdl b/lib/iris/tests/results/integration/netcdf/aux_factories/TestAtmosphereSigma/save.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestAtmosphereSigma/save.cdl
rename to lib/iris/tests/results/integration/netcdf/aux_factories/TestAtmosphereSigma/save.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestHybridPressure/save.cdl b/lib/iris/tests/results/integration/netcdf/aux_factories/TestHybridPressure/save.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestHybridPressure/save.cdl
rename to lib/iris/tests/results/integration/netcdf/aux_factories/TestHybridPressure/save.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestSaveMultipleAuxFactories/hybrid_height_and_pressure.cdl b/lib/iris/tests/results/integration/netcdf/aux_factories/TestSaveMultipleAuxFactories/hybrid_height_and_pressure.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestSaveMultipleAuxFactories/hybrid_height_and_pressure.cdl
rename to lib/iris/tests/results/integration/netcdf/aux_factories/TestSaveMultipleAuxFactories/hybrid_height_and_pressure.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestSaveMultipleAuxFactories/hybrid_height_cubes.cml b/lib/iris/tests/results/integration/netcdf/aux_factories/TestSaveMultipleAuxFactories/hybrid_height_cubes.cml
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestSaveMultipleAuxFactories/hybrid_height_cubes.cml
rename to lib/iris/tests/results/integration/netcdf/aux_factories/TestSaveMultipleAuxFactories/hybrid_height_cubes.cml
diff --git a/lib/iris/tests/results/integration/netcdf/TestPackedData/multi_packed_multi_dtype.cdl b/lib/iris/tests/results/integration/netcdf/general/TestPackedData/multi_packed_multi_dtype.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestPackedData/multi_packed_multi_dtype.cdl
rename to lib/iris/tests/results/integration/netcdf/general/TestPackedData/multi_packed_multi_dtype.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestPackedData/multi_packed_single_dtype.cdl b/lib/iris/tests/results/integration/netcdf/general/TestPackedData/multi_packed_single_dtype.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestPackedData/multi_packed_single_dtype.cdl
rename to lib/iris/tests/results/integration/netcdf/general/TestPackedData/multi_packed_single_dtype.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestPackedData/single_packed_manual.cdl b/lib/iris/tests/results/integration/netcdf/general/TestPackedData/single_packed_manual.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestPackedData/single_packed_manual.cdl
rename to lib/iris/tests/results/integration/netcdf/general/TestPackedData/single_packed_manual.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestPackedData/single_packed_signed.cdl b/lib/iris/tests/results/integration/netcdf/general/TestPackedData/single_packed_signed.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestPackedData/single_packed_signed.cdl
rename to lib/iris/tests/results/integration/netcdf/general/TestPackedData/single_packed_signed.cdl
diff --git a/lib/iris/tests/results/integration/netcdf/TestPackedData/single_packed_unsigned.cdl b/lib/iris/tests/results/integration/netcdf/general/TestPackedData/single_packed_unsigned.cdl
similarity index 100%
rename from lib/iris/tests/results/integration/netcdf/TestPackedData/single_packed_unsigned.cdl
rename to lib/iris/tests/results/integration/netcdf/general/TestPackedData/single_packed_unsigned.cdl
diff --git a/lib/iris/tests/runner/_runner.py b/lib/iris/tests/runner/_runner.py
index bfb2cc2402..7f9439d4b6 100644
--- a/lib/iris/tests/runner/_runner.py
+++ b/lib/iris/tests/runner/_runner.py
@@ -35,18 +35,13 @@ class TestRunner:
("system-tests", "s", "Run the limited subset of system tests."),
("gallery-tests", "e", "Run the gallery code tests."),
("default-tests", "d", "Run the default tests."),
- (
- "coding-tests",
- "c",
- "Run the coding standards tests. (These are a "
- "subset of the default tests.)",
- ),
(
"num-processors=",
"p",
"The number of processors used for running " "the tests.",
),
("create-missing", "m", "Create missing test result files."),
+ ("coverage", "c", "Enable coverage testing"),
]
boolean_options = [
"no-data",
@@ -54,8 +49,8 @@ class TestRunner:
"stop",
"gallery-tests",
"default-tests",
- "coding-tests",
"create-missing",
+ "coverage",
]
def initialize_options(self):
@@ -64,9 +59,9 @@ def initialize_options(self):
self.system_tests = False
self.gallery_tests = False
self.default_tests = False
- self.coding_tests = False
self.num_processors = None
self.create_missing = False
+ self.coverage = False
def finalize_options(self):
# These environment variables will be propagated to all the
@@ -84,8 +79,6 @@ def finalize_options(self):
tests.append("system")
if self.default_tests:
tests.append("default")
- if self.coding_tests:
- tests.append("coding")
if self.gallery_tests:
tests.append("gallery")
if not tests:
@@ -109,8 +102,6 @@ def run(self):
tests.append("lib/iris/tests/system_test.py")
if self.default_tests:
tests.append("lib/iris/tests")
- if self.coding_tests:
- tests.append("lib/iris/tests/test_coding_standards.py")
if self.gallery_tests:
import iris.config
@@ -136,6 +127,9 @@ def run(self):
if self.stop:
args.append("-x")
+ if self.coverage:
+ args.extend(["--cov=lib/iris", "--cov-report=xml"])
+
result = True
for test in tests:
args[0] = test
diff --git a/lib/iris/tests/stock/netcdf.py b/lib/iris/tests/stock/netcdf.py
index e32f065625..bf93f01f6b 100644
--- a/lib/iris/tests/stock/netcdf.py
+++ b/lib/iris/tests/stock/netcdf.py
@@ -12,9 +12,9 @@
import dask
from dask import array as da
-import netCDF4
import numpy as np
+from iris.fileformats.netcdf import _thread_safe_nc
from iris.tests import env_bin_path
NCGEN_PATHSTR = str(env_bin_path("ncgen"))
@@ -100,7 +100,7 @@ def _add_standard_data(nc_path, unlimited_dim_size=0):
"""
- ds = netCDF4.Dataset(nc_path, "r+")
+ ds = _thread_safe_nc.DatasetWrapper(nc_path, "r+")
unlimited_dim_names = [
dim for dim in ds.dimensions if ds.dimensions[dim].isunlimited()
diff --git a/lib/iris/tests/test_aggregate_by.py b/lib/iris/tests/test_aggregate_by.py
index 90bf0e5d4e..e5614f6b63 100644
--- a/lib/iris/tests/test_aggregate_by.py
+++ b/lib/iris/tests/test_aggregate_by.py
@@ -413,6 +413,30 @@ def test_single(self):
aggregateby_cube.data, self.single_rms_expected
)
+ def test_str_aggregation_single_weights_none(self):
+ # mean group-by with single coordinate name.
+ aggregateby_cube = self.cube_single.aggregated_by(
+ "height", iris.analysis.MEAN, weights=None
+ )
+ self.assertCML(
+ aggregateby_cube, ("analysis", "aggregated_by", "single.cml")
+ )
+ np.testing.assert_almost_equal(
+ aggregateby_cube.data, self.single_expected
+ )
+
+ def test_coord_aggregation_single_weights_none(self):
+ # mean group-by with single coordinate.
+ aggregateby_cube = self.cube_single.aggregated_by(
+ self.coord_z_single, iris.analysis.MEAN, weights=None
+ )
+ self.assertCML(
+ aggregateby_cube, ("analysis", "aggregated_by", "single.cml")
+ )
+ np.testing.assert_almost_equal(
+ aggregateby_cube.data, self.single_expected
+ )
+
def test_weighted_single(self):
# weighted mean group-by with single coordinate name.
aggregateby_cube = self.cube_single.aggregated_by(
@@ -1328,5 +1352,153 @@ def test_weights_fail_with_non_weighted_aggregator(self):
)
+# Simply redo the tests of TestAggregateBy with other cubes as weights
+# Note: other weights types (e.g., coordinates, cell measures, etc.) are not
+# tested this way here since this would require adding dimensional metadata
+# objects to the cubes, which would change the CMLs of all resulting cubes of
+# TestAggregateBy.
+
+
+class TestAggregateByWeightedByCube(TestAggregateBy):
+ def setUp(self):
+ super().setUp()
+
+ self.weights_single = self.cube_single[:, 0, 0].copy(
+ self.weights_single
+ )
+ self.weights_single.units = "m2"
+ self.weights_multi = self.cube_multi[:, 0, 0].copy(self.weights_multi)
+ self.weights_multi.units = "m2"
+
+ def test_str_aggregation_weighted_sum_single(self):
+ aggregateby_cube = self.cube_single.aggregated_by(
+ "height",
+ iris.analysis.SUM,
+ weights=self.weights_single,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_coord_aggregation_weighted_sum_single(self):
+ aggregateby_cube = self.cube_single.aggregated_by(
+ self.coord_z_single,
+ iris.analysis.SUM,
+ weights=self.weights_single,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_str_aggregation_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ ["height", "level"],
+ iris.analysis.SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_str_aggregation_rev_order_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ ["level", "height"],
+ iris.analysis.SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_coord_aggregation_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ [self.coord_z1_multi, self.coord_z2_multi],
+ iris.analysis.SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_coord_aggregation_rev_order_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ [self.coord_z2_multi, self.coord_z1_multi],
+ iris.analysis.SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+
+class TestAggregateByWeightedByObj(tests.IrisTest):
+ def setUp(self):
+ self.dim_coord = iris.coords.DimCoord(
+ [0, 1, 2], standard_name="latitude", units="degrees"
+ )
+ self.aux_coord = iris.coords.AuxCoord(
+ [0, 1, 1], long_name="auxcoord", units="kg"
+ )
+ self.cell_measure = iris.coords.CellMeasure(
+ [0, 0, 0], standard_name="cell_area", units="m2"
+ )
+ self.ancillary_variable = iris.coords.AncillaryVariable(
+ [1, 1, 1], var_name="ancvar", units="kg"
+ )
+ self.cube = iris.cube.Cube(
+ [1, 2, 3],
+ standard_name="air_temperature",
+ units="K",
+ dim_coords_and_dims=[(self.dim_coord, 0)],
+ aux_coords_and_dims=[(self.aux_coord, 0)],
+ cell_measures_and_dims=[(self.cell_measure, 0)],
+ ancillary_variables_and_dims=[(self.ancillary_variable, 0)],
+ )
+
+ def test_weighting_with_str_dim_coord(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights="latitude"
+ )
+ np.testing.assert_array_equal(res_cube.data, [0, 8])
+ self.assertEqual(res_cube.units, "K degrees")
+
+ def test_weighting_with_str_aux_coord(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights="auxcoord"
+ )
+ np.testing.assert_array_equal(res_cube.data, [0, 5])
+ self.assertEqual(res_cube.units, "K kg")
+
+ def test_weighting_with_str_cell_measure(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights="cell_area"
+ )
+ np.testing.assert_array_equal(res_cube.data, [0, 0])
+ self.assertEqual(res_cube.units, "K m2")
+
+ def test_weighting_with_str_ancillary_variable(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights="ancvar"
+ )
+ np.testing.assert_array_equal(res_cube.data, [1, 5])
+ self.assertEqual(res_cube.units, "K kg")
+
+ def test_weighting_with_dim_coord(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights=self.dim_coord
+ )
+ np.testing.assert_array_equal(res_cube.data, [0, 8])
+ self.assertEqual(res_cube.units, "K degrees")
+
+ def test_weighting_with_aux_coord(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights=self.aux_coord
+ )
+ np.testing.assert_array_equal(res_cube.data, [0, 5])
+ self.assertEqual(res_cube.units, "K kg")
+
+ def test_weighting_with_cell_measure(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights=self.cell_measure
+ )
+ np.testing.assert_array_equal(res_cube.data, [0, 0])
+ self.assertEqual(res_cube.units, "K m2")
+
+ def test_weighting_with_ancillary_variable(self):
+ res_cube = self.cube.aggregated_by(
+ "auxcoord", iris.analysis.SUM, weights=self.ancillary_variable
+ )
+ np.testing.assert_array_equal(res_cube.data, [1, 5])
+ self.assertEqual(res_cube.units, "K kg")
+
+
if __name__ == "__main__":
unittest.main()
diff --git a/lib/iris/tests/test_analysis.py b/lib/iris/tests/test_analysis.py
index e0a5d0971e..0717368d98 100644
--- a/lib/iris/tests/test_analysis.py
+++ b/lib/iris/tests/test_analysis.py
@@ -12,6 +12,7 @@
import dask.array as da
import numpy as np
import numpy.ma as ma
+import pytest
import iris
import iris.analysis.cartography
@@ -288,7 +289,7 @@ def test_weighted_mean(self):
iris.analysis.MEAN,
)
- # Test collpasing of non data coord
+ # Test collapsing of non data coord
self.assertRaises(
iris.exceptions.CoordinateCollapseError,
e.collapsed,
@@ -1702,5 +1703,237 @@ def test_weights_in_kwargs(self):
self.assertEqual(kwargs, {"test_kwarg": "test", "weights": "ignored"})
+class TestWeights:
+ @pytest.fixture(autouse=True)
+ def setup_test_data(self):
+ self.lat = iris.coords.DimCoord(
+ [0, 1], standard_name="latitude", units="degrees"
+ )
+ self.lon = iris.coords.DimCoord(
+ [0, 1, 2], standard_name="longitude", units="degrees"
+ )
+ self.cell_measure = iris.coords.CellMeasure(
+ np.arange(6).reshape(2, 3), standard_name="cell_area", units="m2"
+ )
+ self.aux_coord = iris.coords.AuxCoord(
+ [3, 4], long_name="auxcoord", units="s"
+ )
+ self.ancillary_variable = iris.coords.AncillaryVariable(
+ [5, 6, 7], var_name="ancvar", units="kg"
+ )
+ self.cube = iris.cube.Cube(
+ np.arange(6).reshape(2, 3),
+ standard_name="air_temperature",
+ units="K",
+ dim_coords_and_dims=[(self.lat, 0), (self.lon, 1)],
+ aux_coords_and_dims=[(self.aux_coord, 0)],
+ cell_measures_and_dims=[(self.cell_measure, (0, 1))],
+ ancillary_variables_and_dims=[(self.ancillary_variable, 1)],
+ )
+
+ def test_init_with_weights(self):
+ weights = iris.analysis._Weights([], self.cube)
+ new_weights = iris.analysis._Weights(weights, self.cube)
+ assert isinstance(new_weights, iris.analysis._Weights)
+ assert new_weights is not weights
+ np.testing.assert_array_equal(new_weights, [])
+ assert new_weights.units == "1"
+ assert weights.units == "1"
+
+ def test_init_with_weights_and_units(self):
+ weights = iris.analysis._Weights([], self.cube)
+ new_weights = iris.analysis._Weights(weights, self.cube, units="J")
+ assert isinstance(new_weights, iris.analysis._Weights)
+ assert new_weights is not weights
+ np.testing.assert_array_equal(new_weights, [])
+ assert new_weights.units == "J"
+ assert weights.units == "1"
+
+ def test_init_with_cube(self):
+ weights = iris.analysis._Weights(self.cube, self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.arange(6).reshape(2, 3))
+ assert weights.units == "K"
+
+ def test_init_with_cube_and_units(self):
+ weights = iris.analysis._Weights(self.cube, self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.arange(6).reshape(2, 3))
+ assert weights.units == "J"
+
+ def test_init_with_str_dim_coord(self):
+ weights = iris.analysis._Weights("latitude", self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[0, 0, 0], [1, 1, 1]])
+ assert weights.units == "degrees"
+
+ def test_init_with_str_dim_coord_and_units(self):
+ weights = iris.analysis._Weights("latitude", self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[0, 0, 0], [1, 1, 1]])
+ assert weights.units == "J"
+
+ def test_init_with_str_aux_coord(self):
+ weights = iris.analysis._Weights("auxcoord", self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[3, 3, 3], [4, 4, 4]])
+ assert weights.units == "s"
+
+ def test_init_with_str_aux_coord_and_units(self):
+ weights = iris.analysis._Weights("auxcoord", self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[3, 3, 3], [4, 4, 4]])
+ assert weights.units == "J"
+
+ def test_init_with_str_ancillary_variable(self):
+ weights = iris.analysis._Weights("ancvar", self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[5, 6, 7], [5, 6, 7]])
+ assert weights.units == "kg"
+
+ def test_init_with_str_ancillary_variable_and_units(self):
+ weights = iris.analysis._Weights("ancvar", self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[5, 6, 7], [5, 6, 7]])
+ assert weights.units == "J"
+
+ def test_init_with_str_cell_measure(self):
+ weights = iris.analysis._Weights("cell_area", self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.arange(6).reshape(2, 3))
+ assert weights.units == "m2"
+
+ def test_init_with_str_cell_measure_and_units(self):
+ weights = iris.analysis._Weights("cell_area", self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.arange(6).reshape(2, 3))
+ assert weights.units == "J"
+
+ def test_init_with_dim_coord(self):
+ weights = iris.analysis._Weights(self.lat, self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[0, 0, 0], [1, 1, 1]])
+ assert weights.units == "degrees"
+
+ def test_init_with_dim_coord_and_units(self):
+ weights = iris.analysis._Weights(self.lat, self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[0, 0, 0], [1, 1, 1]])
+ assert weights.units == "J"
+
+ def test_init_with_aux_coord(self):
+ weights = iris.analysis._Weights(self.aux_coord, self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[3, 3, 3], [4, 4, 4]])
+ assert weights.units == "s"
+
+ def test_init_with_aux_coord_and_units(self):
+ weights = iris.analysis._Weights(self.aux_coord, self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[3, 3, 3], [4, 4, 4]])
+ assert weights.units == "J"
+
+ def test_init_with_ancillary_variable(self):
+ weights = iris.analysis._Weights(self.ancillary_variable, self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[5, 6, 7], [5, 6, 7]])
+ assert weights.units == "kg"
+
+ def test_init_with_ancillary_variable_and_units(self):
+ weights = iris.analysis._Weights(
+ self.ancillary_variable, self.cube, units="J"
+ )
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [[5, 6, 7], [5, 6, 7]])
+ assert weights.units == "J"
+
+ def test_init_with_cell_measure(self):
+ weights = iris.analysis._Weights(self.cell_measure, self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.arange(6).reshape(2, 3))
+ assert weights.units == "m2"
+
+ def test_init_with_cell_measure_and_units(self):
+ weights = iris.analysis._Weights(
+ self.cell_measure, self.cube, units="J"
+ )
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.arange(6).reshape(2, 3))
+ assert weights.units == "J"
+
+ def test_init_with_list(self):
+ weights = iris.analysis._Weights([1, 2, 3], self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [1, 2, 3])
+ assert weights.units == "1"
+
+ def test_init_with_list_and_units(self):
+ weights = iris.analysis._Weights([1, 2, 3], self.cube, units="J")
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, [1, 2, 3])
+ assert weights.units == "J"
+
+ def test_init_with_ndarray(self):
+ weights = iris.analysis._Weights(np.zeros((5, 5)), self.cube)
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.zeros((5, 5)))
+ assert weights.units == "1"
+
+ def test_init_with_ndarray_and_units(self):
+ weights = iris.analysis._Weights(
+ np.zeros((5, 5)), self.cube, units="J"
+ )
+ assert isinstance(weights, iris.analysis._Weights)
+ np.testing.assert_array_equal(weights, np.zeros((5, 5)))
+ assert weights.units == "J"
+
+ def test_init_with_invalid_obj(self):
+ with pytest.raises(KeyError):
+ iris.analysis._Weights("invalid_obj", self.cube)
+
+ def test_init_with_invalid_obj_and_units(self):
+ with pytest.raises(KeyError):
+ iris.analysis._Weights("invalid_obj", self.cube, units="J")
+
+ def test_update_kwargs_no_weights(self):
+ kwargs = {"test": [1, 2, 3]}
+ iris.analysis._Weights.update_kwargs(kwargs, self.cube)
+ assert kwargs == {"test": [1, 2, 3]}
+
+ def test_update_kwargs_weights_none(self):
+ kwargs = {"test": [1, 2, 3], "weights": None}
+ iris.analysis._Weights.update_kwargs(kwargs, self.cube)
+ assert kwargs == {"test": [1, 2, 3], "weights": None}
+
+ def test_update_kwargs_weights(self):
+ kwargs = {"test": [1, 2, 3], "weights": [1, 2]}
+ iris.analysis._Weights.update_kwargs(kwargs, self.cube)
+ assert len(kwargs) == 2
+ assert kwargs["test"] == [1, 2, 3]
+ assert isinstance(kwargs["weights"], iris.analysis._Weights)
+ np.testing.assert_array_equal(kwargs["weights"], [1, 2])
+ assert kwargs["weights"].units == "1"
+
+
+CUBE = iris.cube.Cube(0)
+
+
+@pytest.mark.parametrize(
+ "kwargs,expected",
+ [
+ ({}, "s"),
+ ({"test": "m"}, "s"),
+ ({"weights": None}, "s"),
+ ({"weights": [1, 2, 3]}, "s"),
+ ({"weights": iris.analysis._Weights([1], CUBE)}, "s"),
+ ({"weights": iris.analysis._Weights([1], CUBE, units="kg")}, "s kg"),
+ ],
+)
+def test_sum_units_func(kwargs, expected):
+ units = cf_units.Unit("s")
+ result = iris.analysis._sum_units_func(units, **kwargs)
+ assert result == expected
+
+
if __name__ == "__main__":
tests.main()
diff --git a/lib/iris/tests/test_cdm.py b/lib/iris/tests/test_cdm.py
index 0615dc39bf..8f2a9b474d 100644
--- a/lib/iris/tests/test_cdm.py
+++ b/lib/iris/tests/test_cdm.py
@@ -349,7 +349,6 @@ def test_similar_coord(self):
)
def test_cube_summary_cell_methods(self):
-
cube = self.cube_2d.copy()
# Create a list of values used to create cell methods
diff --git a/lib/iris/tests/test_cf.py b/lib/iris/tests/test_cf.py
index 034fb1dbda..3abd6b981b 100644
--- a/lib/iris/tests/test_cf.py
+++ b/lib/iris/tests/test_cf.py
@@ -15,6 +15,8 @@
import io
from unittest import mock
+import pytest
+
import iris
import iris.fileformats.cf as cf
@@ -52,11 +54,14 @@ def test_cached(self):
@tests.skip_data
class TestCFReader(tests.IrisTest):
- def setUp(self):
+ @pytest.fixture(autouse=True)
+ def set_up(self):
filename = tests.get_data_path(
("NetCDF", "rotated", "xyt", "small_rotPole_precipitation.nc")
)
self.cfr = cf.CFReader(filename)
+ with self.cfr:
+ yield
def test_ancillary_variables_pass_0(self):
self.assertEqual(self.cfr.cf_group.ancillary_variables, {})
@@ -276,9 +281,7 @@ def test_destructor(self):
didn't exist because opening the dataset had failed.
"""
with self.temp_filename(suffix=".nc") as fn:
-
with open(fn, "wb+") as fh:
-
fh.write(
b"\x89HDF\r\n\x1a\nBroken file with correct signature"
)
@@ -350,7 +353,8 @@ def test_cell_methods(self):
@tests.skip_data
class TestClimatology(tests.IrisTest):
- def setUp(self):
+ @pytest.fixture(autouse=True)
+ def set_up(self):
filename = tests.get_data_path(
(
"NetCDF",
@@ -359,6 +363,8 @@ def setUp(self):
)
)
self.cfr = cf.CFReader(filename)
+ with self.cfr:
+ yield
def test_bounds(self):
time = self.cfr.cf_group["temp_dmax_tmean_abs"].cf_group.coordinates[
@@ -375,7 +381,8 @@ def test_bounds(self):
@tests.skip_data
class TestLabels(tests.IrisTest):
- def setUp(self):
+ @pytest.fixture(autouse=True)
+ def set_up(self):
filename = tests.get_data_path(
(
"NetCDF",
@@ -390,6 +397,10 @@ def setUp(self):
)
self.cfr_end = cf.CFReader(filename)
+ with self.cfr_start:
+ with self.cfr_end:
+ yield
+
def test_label_dim_start(self):
cf_data_var = self.cfr_start.cf_group["temp_dmax_tmean_abs"]
diff --git a/lib/iris/tests/test_coding_standards.py b/lib/iris/tests/test_coding_standards.py
index 01f6f777fa..b52934c568 100644
--- a/lib/iris/tests/test_coding_standards.py
+++ b/lib/iris/tests/test_coding_standards.py
@@ -12,9 +12,12 @@
from fnmatch import fnmatch
from glob import glob
import os
+from pathlib import Path
import subprocess
import iris
+from iris.fileformats.netcdf import _thread_safe_nc
+from iris.tests import system_test
LICENSE_TEMPLATE = """# Copyright Iris contributors
#
@@ -40,6 +43,29 @@
IRIS_REPO_DIRPATH = os.environ.get("IRIS_REPO_DIR", IRIS_INSTALL_DIR)
+def test_netcdf4_import():
+ """Use of netCDF4 must be via iris.fileformats.netcdf._thread_safe_nc ."""
+ # Please avoid including these phrases in any comments/strings throughout
+ # Iris (e.g. use "from the netCDF4 library" instead) - this allows the
+ # below search to remain quick and simple.
+ import_strings = ("import netCDF4", "from netCDF4")
+
+ files_including_import = []
+ for file_path in Path(IRIS_DIR).rglob("*.py"):
+ with file_path.open("r") as open_file:
+ file_text = open_file.read()
+
+ if any([i in file_text for i in import_strings]):
+ files_including_import.append(file_path)
+
+ expected = [
+ Path(_thread_safe_nc.__file__),
+ Path(system_test.__file__),
+ Path(__file__),
+ ]
+ assert set(files_including_import) == set(expected)
+
+
class TestLicenseHeaders(tests.IrisTest):
@staticmethod
def whatchanged_parse(whatchanged_output):
diff --git a/lib/iris/tests/test_io_init.py b/lib/iris/tests/test_io_init.py
index d33b76ddeb..82da82cfa9 100644
--- a/lib/iris/tests/test_io_init.py
+++ b/lib/iris/tests/test_io_init.py
@@ -126,7 +126,7 @@ def test_format_picker(self):
]
# test that each filespec is identified as the expected format
- for (expected_format_name, file_spec) in test_specs:
+ for expected_format_name, file_spec in test_specs:
test_path = tests.get_data_path(file_spec)
with open(test_path, "rb") as test_file:
a = iff.FORMAT_AGENT.get_spec(test_path, test_file)
diff --git a/lib/iris/tests/test_lazy_aggregate_by.py b/lib/iris/tests/test_lazy_aggregate_by.py
index d1ebc9a36a..57b748e52f 100644
--- a/lib/iris/tests/test_lazy_aggregate_by.py
+++ b/lib/iris/tests/test_lazy_aggregate_by.py
@@ -6,6 +6,7 @@
import unittest
from iris._lazy_data import as_lazy_data
+from iris.analysis import SUM
from iris.tests import test_aggregate_by
@@ -44,5 +45,65 @@ def tearDown(self):
assert self.cube_easy.has_lazy_data()
+class TestLazyAggregateByWeightedByCube(TestLazyAggregateBy):
+ def setUp(self):
+ super().setUp()
+
+ self.weights_single = self.cube_single[:, 0, 0].copy(
+ self.weights_single
+ )
+ self.weights_single.units = "m2"
+ self.weights_multi = self.cube_multi[:, 0, 0].copy(self.weights_multi)
+ self.weights_multi.units = "m2"
+
+ def test_str_aggregation_weighted_sum_single(self):
+ aggregateby_cube = self.cube_single.aggregated_by(
+ "height",
+ SUM,
+ weights=self.weights_single,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_coord_aggregation_weighted_sum_single(self):
+ aggregateby_cube = self.cube_single.aggregated_by(
+ self.coord_z_single,
+ SUM,
+ weights=self.weights_single,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_str_aggregation_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ ["height", "level"],
+ SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_str_aggregation_rev_order_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ ["level", "height"],
+ SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_coord_aggregation_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ [self.coord_z1_multi, self.coord_z2_multi],
+ SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+ def test_coord_aggregation_rev_order_weighted_sum_multi(self):
+ aggregateby_cube = self.cube_multi.aggregated_by(
+ [self.coord_z2_multi, self.coord_z1_multi],
+ SUM,
+ weights=self.weights_multi,
+ )
+ self.assertEqual(aggregateby_cube.units, "kelvin m2")
+
+
if __name__ == "__main__":
unittest.main()
diff --git a/lib/iris/tests/test_load.py b/lib/iris/tests/test_load.py
index 4749236abc..adb33924e5 100644
--- a/lib/iris/tests/test_load.py
+++ b/lib/iris/tests/test_load.py
@@ -14,9 +14,8 @@
import pathlib
from unittest import mock
-import netCDF4
-
import iris
+from iris.fileformats.netcdf import _thread_safe_nc
import iris.io
@@ -193,10 +192,11 @@ def test_netCDF_Dataset_call(self):
filename = tests.get_data_path(
("NetCDF", "global", "xyt", "SMALL_total_column_co2.nc")
)
- fake_dataset = netCDF4.Dataset(filename)
+ fake_dataset = _thread_safe_nc.DatasetWrapper(filename)
with mock.patch(
- "netCDF4.Dataset", return_value=fake_dataset
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=fake_dataset,
) as dataset_loader:
next(iris.io.load_http([self.url], callback=None))
dataset_loader.assert_called_with(self.url, mode="r")
diff --git a/lib/iris/tests/test_merge.py b/lib/iris/tests/test_merge.py
index c209d68da0..e53bbfb5f3 100644
--- a/lib/iris/tests/test_merge.py
+++ b/lib/iris/tests/test_merge.py
@@ -190,7 +190,7 @@ def setUp(self):
)
def test__ndarray_ndarray(self):
- for (lazy0, lazy1) in self.lazy_combos:
+ for lazy0, lazy1 in self.lazy_combos:
cubes = iris.cube.CubeList()
cubes.append(self._make_cube(0, dtype=self.dtype, lazy=lazy0))
cubes.append(self._make_cube(1, dtype=self.dtype, lazy=lazy1))
diff --git a/lib/iris/tests/test_netcdf.py b/lib/iris/tests/test_netcdf.py
index 5017698a22..d182de84f6 100644
--- a/lib/iris/tests/test_netcdf.py
+++ b/lib/iris/tests/test_netcdf.py
@@ -19,7 +19,6 @@
import tempfile
from unittest import mock
-import netCDF4 as nc
import numpy as np
import numpy.ma as ma
@@ -29,6 +28,7 @@
import iris.coord_systems as icoord_systems
from iris.fileformats._nc_load_rules import helpers as ncload_helpers
import iris.fileformats.netcdf
+from iris.fileformats.netcdf import _thread_safe_nc
from iris.fileformats.netcdf import load_cubes as nc_load_cubes
import iris.std_names
import iris.tests.stock as stock
@@ -81,7 +81,7 @@ def test_missing_time_bounds(self):
("NetCDF", "global", "xyt", "SMALL_hires_wind_u_for_ipcc4.nc")
)
shutil.copyfile(src, filename)
- dataset = nc.Dataset(filename, mode="a")
+ dataset = _thread_safe_nc.DatasetWrapper(filename, mode="a")
dataset.renameVariable("time_bnds", "foo")
dataset.close()
_ = iris.load_cube(filename, "eastward_wind")
@@ -204,7 +204,7 @@ def test_missing_climatology(self):
("NetCDF", "transverse_mercator", "tmean_1910_1910.nc")
)
shutil.copyfile(src, filename)
- dataset = nc.Dataset(filename, mode="a")
+ dataset = _thread_safe_nc.DatasetWrapper(filename, mode="a")
dataset.renameVariable("climatology_bounds", "foo")
dataset.close()
_ = iris.load_cube(filename, "Mean temperature")
@@ -313,9 +313,7 @@ def test_deferred_loading(self):
cube[((0, 8, 4, 2, 14, 12),)][((0, 2, 4, 1),)],
("netcdf", "netcdf_deferred_tuple_1.cml"),
)
- subcube = cube[((0, 8, 4, 2, 14, 12),)][((0, 2, 4, 1),)][
- (1, 3),
- ]
+ subcube = cube[((0, 8, 4, 2, 14, 12),)][((0, 2, 4, 1),)][(1, 3),]
self.assertCML(subcube, ("netcdf", "netcdf_deferred_tuple_2.cml"))
# Consecutive mixture on same dimension.
@@ -634,7 +632,7 @@ def test_netcdf_save_format(self):
with self.temp_filename(suffix=".nc") as file_out:
# Test default NETCDF4 file format saving.
iris.save(cube, file_out)
- ds = nc.Dataset(file_out)
+ ds = _thread_safe_nc.DatasetWrapper(file_out)
self.assertEqual(
ds.file_format, "NETCDF4", "Failed to save as NETCDF4 format"
)
@@ -642,7 +640,7 @@ def test_netcdf_save_format(self):
# Test NETCDF4_CLASSIC file format saving.
iris.save(cube, file_out, netcdf_format="NETCDF4_CLASSIC")
- ds = nc.Dataset(file_out)
+ ds = _thread_safe_nc.DatasetWrapper(file_out)
self.assertEqual(
ds.file_format,
"NETCDF4_CLASSIC",
@@ -652,7 +650,7 @@ def test_netcdf_save_format(self):
# Test NETCDF3_CLASSIC file format saving.
iris.save(cube, file_out, netcdf_format="NETCDF3_CLASSIC")
- ds = nc.Dataset(file_out)
+ ds = _thread_safe_nc.DatasetWrapper(file_out)
self.assertEqual(
ds.file_format,
"NETCDF3_CLASSIC",
@@ -662,7 +660,7 @@ def test_netcdf_save_format(self):
# Test NETCDF4_64BIT file format saving.
iris.save(cube, file_out, netcdf_format="NETCDF3_64BIT")
- ds = nc.Dataset(file_out)
+ ds = _thread_safe_nc.DatasetWrapper(file_out)
self.assertTrue(
ds.file_format in ["NETCDF3_64BIT", "NETCDF3_64BIT_OFFSET"],
"Failed to save as NETCDF3_64BIT format",
@@ -1049,7 +1047,7 @@ def test_attributes(self):
with self.temp_filename(suffix=".nc") as filename:
iris.save(self.cube, filename)
# Load the dataset.
- ds = nc.Dataset(filename, "r")
+ ds = _thread_safe_nc.DatasetWrapper(filename, "r")
exceptions = []
# Should be global attributes.
for gkey in aglobals:
@@ -1213,7 +1211,7 @@ def test_shared(self):
self.assertCDL(filename)
# Also check that only one, shared ancillary variable was written.
- ds = nc.Dataset(filename)
+ ds = _thread_safe_nc.DatasetWrapper(filename)
self.assertIn("air_potential_temperature", ds.variables)
self.assertIn("alternate_data", ds.variables)
self.assertEqual(
@@ -1417,7 +1415,6 @@ def test_process_flags(self):
}
for bits, descriptions in multiple_map.items():
-
ll_cube = stock.lat_lon_cube()
ll_cube.attributes["ukmo__process_flags"] = descriptions
diff --git a/lib/iris/tests/test_nimrod.py b/lib/iris/tests/test_nimrod.py
index a1d7bb298f..6d62623198 100644
--- a/lib/iris/tests/test_nimrod.py
+++ b/lib/iris/tests/test_nimrod.py
@@ -80,7 +80,8 @@ def test_huge_field_load(self):
@tests.skip_data
def test_load_kwarg(self):
"""Tests that the handle_metadata_errors kwarg is effective by setting it to
- False with a file with known incomplete meta-data (missing ellipsoid)."""
+ False with a file with known incomplete meta-data (missing ellipsoid).
+ """
datafile = "u1096_ng_ek00_pressure_2km"
with self.assertRaisesRegex(
TranslationError,
diff --git a/lib/iris/tests/test_pp_cf.py b/lib/iris/tests/test_pp_cf.py
index 2b497cb53b..49bedaf1e2 100644
--- a/lib/iris/tests/test_pp_cf.py
+++ b/lib/iris/tests/test_pp_cf.py
@@ -10,10 +10,9 @@
import os
import tempfile
-import netCDF4
-
import iris
import iris.coords
+from iris.fileformats.netcdf import _thread_safe_nc
from iris.fileformats.pp import STASH
import iris.tests.pp as pp
import iris.util
@@ -95,7 +94,7 @@ def _test_file(self, name):
for index, cube in enumerate(cubes):
# Explicitly set a fill-value as a workaround for
# https://github.com/Unidata/netcdf4-python/issues/725
- fill_value = netCDF4.default_fillvals[cube.dtype.str[1:]]
+ fill_value = _thread_safe_nc.default_fillvals[cube.dtype.str[1:]]
file_nc = tempfile.NamedTemporaryFile(
suffix=".nc", delete=False
diff --git a/lib/iris/tests/test_pp_stash.py b/lib/iris/tests/test_pp_stash.py
index b153aef0d4..42390ab2b3 100644
--- a/lib/iris/tests/test_pp_stash.py
+++ b/lib/iris/tests/test_pp_stash.py
@@ -86,7 +86,6 @@ def test_irregular_stash_str(self):
)
def test_illegal_stash_str_range(self):
-
self.assertEqual(iris.fileformats.pp.STASH(0, 2, 3), "m??s02i003")
self.assertNotEqual(iris.fileformats.pp.STASH(0, 2, 3), "m01s02i003")
@@ -124,7 +123,7 @@ def test_illegal_stash_format(self):
("m01s02003", (1, 2, 3)),
)
- for (test_value, reference) in test_values:
+ for test_value, reference in test_values:
msg = "Expected STASH code .* {!r}".format(test_value)
with self.assertRaisesRegex(ValueError, msg):
test_value == iris.fileformats.pp.STASH(*reference)
@@ -137,7 +136,7 @@ def test_illegal_stash_type(self):
(["m01s02i003"], "m01s02i003"),
)
- for (test_value, reference) in test_values:
+ for test_value, reference in test_values:
msg = "Expected STASH code .* {!r}".format(test_value)
with self.assertRaisesRegex(TypeError, msg):
iris.fileformats.pp.STASH.from_msi(test_value) == reference
diff --git a/lib/iris/tests/test_util.py b/lib/iris/tests/test_util.py
index db182ae3f3..d8d5d73e95 100644
--- a/lib/iris/tests/test_util.py
+++ b/lib/iris/tests/test_util.py
@@ -161,7 +161,6 @@ def test_default_values(self):
)
def test_trim_string_with_no_spaces(self):
-
clip_length = 200
no_space_string = "a" * 500
diff --git a/lib/iris/tests/unit/analysis/cartography/test_gridcell_angles.py b/lib/iris/tests/unit/analysis/cartography/test_gridcell_angles.py
index 6b957baec6..810851362e 100644
--- a/lib/iris/tests/unit/analysis/cartography/test_gridcell_angles.py
+++ b/lib/iris/tests/unit/analysis/cartography/test_gridcell_angles.py
@@ -94,7 +94,6 @@ def _check_multiple_orientations_and_latitudes(
atol_degrees=0.005,
cellsize_degrees=1.0,
):
-
cube = _2d_multicells_testcube(cellsize_degrees=cellsize_degrees)
# Calculate gridcell angles at each point.
diff --git a/lib/iris/tests/unit/analysis/maths/__init__.py b/lib/iris/tests/unit/analysis/maths/__init__.py
index 311da8a0e6..558a6fccfe 100644
--- a/lib/iris/tests/unit/analysis/maths/__init__.py
+++ b/lib/iris/tests/unit/analysis/maths/__init__.py
@@ -247,7 +247,7 @@ def test_partial_mask_second_lazy_not_in_place(self):
def test_in_place_introduces_mask(self):
# If second cube is masked, result should also be masked.
- data1 = np.arange(4, dtype=np.float)
+ data1 = np.arange(4, dtype=float)
data2 = ma.array([2.0, 2.0, 2.0, 2.0], mask=[1, 1, 0, 0])
cube1 = Cube(data1)
cube2 = Cube(data2)
diff --git a/lib/iris/tests/unit/analysis/test_Aggregator.py b/lib/iris/tests/unit/analysis/test_Aggregator.py
index ec837ea49a..45081ad07f 100644
--- a/lib/iris/tests/unit/analysis/test_Aggregator.py
+++ b/lib/iris/tests/unit/analysis/test_Aggregator.py
@@ -15,6 +15,7 @@
import numpy.ma as ma
from iris.analysis import Aggregator
+from iris.cube import Cube
from iris.exceptions import LazyAggregatorError
@@ -286,10 +287,30 @@ def test_units_change(self):
units_func = mock.Mock(return_value=mock.sentinel.new_units)
aggregator = Aggregator("", None, units_func)
cube = mock.Mock(units=mock.sentinel.units)
- aggregator.update_metadata(cube, [])
- units_func.assert_called_once_with(mock.sentinel.units)
+ aggregator.update_metadata(cube, [], kw1=1, kw2=2)
+ units_func.assert_called_once_with(mock.sentinel.units, kw1=1, kw2=2)
self.assertEqual(cube.units, mock.sentinel.new_units)
+ def test_units_func_no_kwargs(self):
+ # To ensure backwards-compatibility, Aggregator also supports
+ # units_func that accept the single argument `units`
+ def units_func(units):
+ return units**2
+
+ aggregator = Aggregator("", None, units_func)
+ cube = Cube(0, units="s")
+ aggregator.update_metadata(cube, [], kw1=1, kw2=2)
+ self.assertEqual(cube.units, "s2")
+
+ def test_units_func_kwargs(self):
+ def units_func(units, **kwargs):
+ return units**2
+
+ aggregator = Aggregator("", None, units_func)
+ cube = Cube(0, units="s")
+ aggregator.update_metadata(cube, [], kw1=1, kw2=2)
+ self.assertEqual(cube.units, "s2")
+
class Test_lazy_aggregate(tests.IrisTest):
def test_kwarg_pass_through_no_kwargs(self):
diff --git a/lib/iris/tests/unit/analysis/test_RMS.py b/lib/iris/tests/unit/analysis/test_RMS.py
index 141b3e262b..74f309ce00 100644
--- a/lib/iris/tests/unit/analysis/test_RMS.py
+++ b/lib/iris/tests/unit/analysis/test_RMS.py
@@ -101,20 +101,16 @@ def test_1d_weighted(self):
data = as_lazy_data(np.array([4, 7, 10, 8], dtype=np.float64))
weights = np.array([1, 4, 3, 2], dtype=np.float64)
expected_rms = 8.0
- # https://github.com/dask/dask/issues/3846.
- with self.assertRaisesRegex(TypeError, "unexpected keyword argument"):
- rms = RMS.lazy_aggregate(data, 0, weights=weights)
- self.assertAlmostEqual(rms, expected_rms)
+ rms = RMS.lazy_aggregate(data, 0, weights=weights)
+ self.assertAlmostEqual(rms, expected_rms)
def test_1d_lazy_weighted(self):
# 1-dimensional input with lazy weights.
data = as_lazy_data(np.array([4, 7, 10, 8], dtype=np.float64))
weights = as_lazy_data(np.array([1, 4, 3, 2], dtype=np.float64))
expected_rms = 8.0
- # https://github.com/dask/dask/issues/3846.
- with self.assertRaisesRegex(TypeError, "unexpected keyword argument"):
- rms = RMS.lazy_aggregate(data, 0, weights=weights)
- self.assertAlmostEqual(rms, expected_rms)
+ rms = RMS.lazy_aggregate(data, 0, weights=weights)
+ self.assertAlmostEqual(rms, expected_rms)
def test_2d_weighted(self):
# 2-dimensional input with weights.
@@ -123,20 +119,16 @@ def test_2d_weighted(self):
)
weights = np.array([[1, 4, 3, 2], [2, 1, 1.5, 0.5]], dtype=np.float64)
expected_rms = np.array([8.0, 16.0], dtype=np.float64)
- # https://github.com/dask/dask/issues/3846.
- with self.assertRaisesRegex(TypeError, "unexpected keyword argument"):
- rms = RMS.lazy_aggregate(data, 1, weights=weights)
- self.assertArrayAlmostEqual(rms, expected_rms)
+ rms = RMS.lazy_aggregate(data, 1, weights=weights)
+ self.assertArrayAlmostEqual(rms, expected_rms)
def test_unit_weighted(self):
# Unit weights should be the same as no weights.
data = as_lazy_data(np.array([5, 2, 6, 4], dtype=np.float64))
weights = np.ones_like(data)
expected_rms = 4.5
- # https://github.com/dask/dask/issues/3846.
- with self.assertRaisesRegex(TypeError, "unexpected keyword argument"):
- rms = RMS.lazy_aggregate(data, 0, weights=weights)
- self.assertAlmostEqual(rms, expected_rms)
+ rms = RMS.lazy_aggregate(data, 0, weights=weights)
+ self.assertAlmostEqual(rms, expected_rms)
def test_masked(self):
# Masked entries should be completely ignored.
@@ -152,9 +144,6 @@ def test_masked(self):
self.assertAlmostEqual(rms, expected_rms)
def test_masked_weighted(self):
- # Weights should work properly with masked arrays, but currently don't
- # (see https://github.com/dask/dask/issues/3846).
- # For now, masked weights are simply not supported.
data = as_lazy_data(
ma.array(
[4, 7, 18, 10, 11, 8],
@@ -164,9 +153,8 @@ def test_masked_weighted(self):
)
weights = np.array([1, 4, 5, 3, 8, 2])
expected_rms = 8.0
- with self.assertRaisesRegex(TypeError, "unexpected keyword argument"):
- rms = RMS.lazy_aggregate(data, 0, weights=weights)
- self.assertAlmostEqual(rms, expected_rms)
+ rms = RMS.lazy_aggregate(data, 0, weights=weights)
+ self.assertAlmostEqual(rms, expected_rms)
class Test_name(tests.IrisTest):
diff --git a/lib/iris/tests/unit/coords/test_AncillaryVariable.py b/lib/iris/tests/unit/coords/test_AncillaryVariable.py
index 75b6250449..e5fc8fd28a 100644
--- a/lib/iris/tests/unit/coords/test_AncillaryVariable.py
+++ b/lib/iris/tests/unit/coords/test_AncillaryVariable.py
@@ -68,7 +68,7 @@ def setUp(self):
self.setupTestArrays(masked=True)
def test_lazyness_and_dtype_combinations(self):
- for (ancill_var, data_lazyness) in data_all_dtypes_and_lazynesses(
+ for ancill_var, data_lazyness in data_all_dtypes_and_lazynesses(
self,
):
data = ancill_var.core_data()
@@ -225,10 +225,9 @@ def test_dtypes(self):
# floating dtype.
# Check that dtypes remain the same in all cases, taking the dtypes
# directly from the core data as we have no masking).
- for (main_ancill_var, data_lazyness) in data_all_dtypes_and_lazynesses(
+ for main_ancill_var, data_lazyness in data_all_dtypes_and_lazynesses(
self
):
-
sub_ancill_var = main_ancill_var[:2, 1]
ancill_var_dtype = main_ancill_var.dtype
@@ -250,10 +249,9 @@ def test_lazyness(self):
# Index ancillary variables with real+lazy data, and either an int or
# floating dtype.
# Check that lazy data stays lazy and real stays real, in all cases.
- for (main_ancill_var, data_lazyness) in data_all_dtypes_and_lazynesses(
+ for main_ancill_var, data_lazyness in data_all_dtypes_and_lazynesses(
self
):
-
sub_ancill_var = main_ancill_var[:2, 1]
msg = (
@@ -277,10 +275,9 @@ def test_lazyness(self):
def test_real_data_copies(self):
# Index ancillary variables with real+lazy data.
# In all cases, check that any real arrays are copied by the indexing.
- for (main_ancill_var, data_lazyness) in data_all_dtypes_and_lazynesses(
+ for main_ancill_var, data_lazyness in data_all_dtypes_and_lazynesses(
self
):
-
sub_ancill_var = main_ancill_var[:2, 1]
msg = (
@@ -308,10 +305,9 @@ def test_lazyness(self):
# Copy ancillary variables with real+lazy data, and either an int or
# floating dtype.
# Check that lazy data stays lazy and real stays real, in all cases.
- for (main_ancill_var, data_lazyness) in data_all_dtypes_and_lazynesses(
+ for main_ancill_var, data_lazyness in data_all_dtypes_and_lazynesses(
self
):
-
ancill_var_dtype = main_ancill_var.dtype
copied_ancill_var = main_ancill_var.copy()
@@ -338,10 +334,9 @@ def test_lazyness(self):
def test_realdata_copies(self):
# Copy ancillary variables with real+lazy data.
# In all cases, check that any real arrays are copies, not views.
- for (main_ancill_var, data_lazyness) in data_all_dtypes_and_lazynesses(
+ for main_ancill_var, data_lazyness in data_all_dtypes_and_lazynesses(
self
):
-
copied_ancill_var = main_ancill_var.copy()
msg = (
@@ -520,79 +515,79 @@ def _check(self, result_ancill_var, expected_data, lazyness):
self.assertEqualLazyArraysAndDtypes(expected_data, data)
def test_add(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = ancill_var + 10
expected_data = orig_data + 10
self._check(result, expected_data, data_lazyness)
def test_add_inplace(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
ancill_var += 10
expected_data = orig_data + 10
self._check(ancill_var, expected_data, data_lazyness)
def test_right_add(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = 10 + ancill_var
expected_data = 10 + orig_data
self._check(result, expected_data, data_lazyness)
def test_subtract(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = ancill_var - 10
expected_data = orig_data - 10
self._check(result, expected_data, data_lazyness)
def test_subtract_inplace(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
ancill_var -= 10
expected_data = orig_data - 10
self._check(ancill_var, expected_data, data_lazyness)
def test_right_subtract(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = 10 - ancill_var
expected_data = 10 - orig_data
self._check(result, expected_data, data_lazyness)
def test_multiply(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = ancill_var * 10
expected_data = orig_data * 10
self._check(result, expected_data, data_lazyness)
def test_multiply_inplace(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
ancill_var *= 10
expected_data = orig_data * 10
self._check(ancill_var, expected_data, data_lazyness)
def test_right_multiply(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = 10 * ancill_var
expected_data = 10 * orig_data
self._check(result, expected_data, data_lazyness)
def test_divide(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = ancill_var / 10
expected_data = orig_data / 10
self._check(result, expected_data, data_lazyness)
def test_divide_inplace(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
ancill_var /= 10
expected_data = orig_data / 10
self._check(ancill_var, expected_data, data_lazyness)
def test_right_divide(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = 10 / ancill_var
expected_data = 10 / orig_data
self._check(result, expected_data, data_lazyness)
def test_negative(self):
- for (ancill_var, orig_data, data_lazyness) in self.test_combinations:
+ for ancill_var, orig_data, data_lazyness in self.test_combinations:
result = -ancill_var
expected_data = -orig_data
self._check(result, expected_data, data_lazyness)
diff --git a/lib/iris/tests/unit/coords/test_AuxCoord.py b/lib/iris/tests/unit/coords/test_AuxCoord.py
index e6cd8ac821..e5147659fc 100644
--- a/lib/iris/tests/unit/coords/test_AuxCoord.py
+++ b/lib/iris/tests/unit/coords/test_AuxCoord.py
@@ -370,7 +370,6 @@ def test_dtypes(self):
points_type_name,
bounds_type_name,
) in coords_all_dtypes_and_lazynesses(self, AuxCoord):
-
sub_coord = main_coord[:2, 1]
coord_dtype = main_coord.dtype
@@ -417,7 +416,6 @@ def test_lazyness(self):
points_type_name,
bounds_type_name,
) in coords_all_dtypes_and_lazynesses(self, AuxCoord):
-
sub_coord = main_coord[:2, 1]
msg = (
@@ -463,7 +461,6 @@ def test_real_data_copies(self):
points_lazyness,
bounds_lazyness,
) in coords_all_dtypes_and_lazynesses(self, AuxCoord):
-
sub_coord = main_coord[:2, 1]
msg = (
@@ -511,7 +508,6 @@ def test_lazyness(self):
points_lazyness,
bounds_lazyness,
) in coords_all_dtypes_and_lazynesses(self, AuxCoord):
-
coord_dtype = main_coord.dtype
copied_coord = main_coord.copy()
@@ -558,7 +554,6 @@ def test_realdata_copies(self):
points_lazyness,
bounds_lazyness,
) in coords_all_dtypes_and_lazynesses(self, AuxCoord):
-
copied_coord = main_coord.copy()
msg = (
diff --git a/lib/iris/tests/unit/coords/test_Coord.py b/lib/iris/tests/unit/coords/test_Coord.py
index dca6ed3c1b..72a48437ec 100644
--- a/lib/iris/tests/unit/coords/test_Coord.py
+++ b/lib/iris/tests/unit/coords/test_Coord.py
@@ -463,7 +463,6 @@ def test_lazy_nd_bounds_last(self):
)
def test_lazy_nd_points_and_bounds(self):
-
self.setupTestArrays((3, 4))
coord = AuxCoord(self.pts_lazy, bounds=self.bds_lazy)
@@ -520,7 +519,6 @@ def test_lazy_nd_noncontiguous_bounds_warning(self):
coord.collapsed()
def test_numeric_3_bounds(self):
-
points = np.array([2.0, 6.0, 4.0])
bounds = np.array([[1.0, 0.0, 3.0], [5.0, 4.0, 7.0], [3.0, 2.0, 5.0]])
@@ -544,7 +542,6 @@ def test_numeric_3_bounds(self):
)
def test_lazy_3_bounds(self):
-
points = da.arange(3) * 2.0
bounds = da.arange(3 * 3).reshape(3, 3)
diff --git a/lib/iris/tests/unit/coords/test_DimCoord.py b/lib/iris/tests/unit/coords/test_DimCoord.py
index 4298b140ea..dd0ba48f3d 100644
--- a/lib/iris/tests/unit/coords/test_DimCoord.py
+++ b/lib/iris/tests/unit/coords/test_DimCoord.py
@@ -304,7 +304,6 @@ def test_dtypes(self):
points_type_name,
bounds_type_name,
) in coords_all_dtypes_and_lazynesses(self, DimCoord):
-
sub_coord = main_coord[:2]
coord_dtype = main_coord.dtype
@@ -404,7 +403,6 @@ def test_real_data_copies(self):
points_lazyness,
bounds_lazyness,
) in coords_all_dtypes_and_lazynesses(self, DimCoord):
-
sub_coord = main_coord[:2]
msg = (
@@ -470,7 +468,6 @@ def test_realdata_readonly(self):
points_type_name,
bounds_type_name,
) in coords_all_dtypes_and_lazynesses(self, DimCoord):
-
copied_coord = main_coord.copy()
copied_points = copied_coord.core_points()
diff --git a/lib/iris/tests/unit/cube/test_Cube.py b/lib/iris/tests/unit/cube/test_Cube.py
index 5d120a6982..aa9e3b51b1 100644
--- a/lib/iris/tests/unit/cube/test_Cube.py
+++ b/lib/iris/tests/unit/cube/test_Cube.py
@@ -20,7 +20,7 @@
from iris._lazy_data import as_lazy_data
import iris.analysis
-from iris.analysis import MEAN, Aggregator, WeightedAggregator
+from iris.analysis import MEAN, SUM, Aggregator, WeightedAggregator
import iris.aux_factory
from iris.aux_factory import HybridHeightFactory
from iris.common.metadata import BaseMetadata
@@ -320,18 +320,36 @@ def test_dim0_lazy(self):
self.assertArrayAlmostEqual(cube_collapsed.data, [1.5, 2.5, 3.5])
self.assertFalse(cube_collapsed.has_lazy_data())
+ def test_dim0_lazy_weights_none(self):
+ cube_collapsed = self.cube.collapsed("y", MEAN, weights=None)
+ self.assertTrue(cube_collapsed.has_lazy_data())
+ self.assertArrayAlmostEqual(cube_collapsed.data, [1.5, 2.5, 3.5])
+ self.assertFalse(cube_collapsed.has_lazy_data())
+
def test_dim1_lazy(self):
cube_collapsed = self.cube.collapsed("x", MEAN)
self.assertTrue(cube_collapsed.has_lazy_data())
self.assertArrayAlmostEqual(cube_collapsed.data, [1.0, 4.0])
self.assertFalse(cube_collapsed.has_lazy_data())
+ def test_dim1_lazy_weights_none(self):
+ cube_collapsed = self.cube.collapsed("x", MEAN, weights=None)
+ self.assertTrue(cube_collapsed.has_lazy_data())
+ self.assertArrayAlmostEqual(cube_collapsed.data, [1.0, 4.0])
+ self.assertFalse(cube_collapsed.has_lazy_data())
+
def test_multidims(self):
# Check that MEAN works with multiple dims.
cube_collapsed = self.cube.collapsed(("x", "y"), MEAN)
self.assertTrue(cube_collapsed.has_lazy_data())
self.assertArrayAllClose(cube_collapsed.data, 2.5)
+ def test_multidims_weights_none(self):
+ # Check that MEAN works with multiple dims.
+ cube_collapsed = self.cube.collapsed(("x", "y"), MEAN, weights=None)
+ self.assertTrue(cube_collapsed.has_lazy_data())
+ self.assertArrayAllClose(cube_collapsed.data, 2.5)
+
def test_non_lazy_aggregator(self):
# An aggregator which doesn't have a lazy function should still work.
dummy_agg = Aggregator(
@@ -342,18 +360,19 @@ def test_non_lazy_aggregator(self):
self.assertArrayEqual(result.data, np.mean(self.data, axis=1))
-class Test_collapsed__multidim_weighted(tests.IrisTest):
+class Test_collapsed__multidim_weighted_with_arr(tests.IrisTest):
def setUp(self):
self.data = np.arange(6.0).reshape((2, 3))
self.lazydata = as_lazy_data(self.data)
- # Test cubes wth (same-valued) real and lazy data
- cube_real = Cube(self.data)
+ # Test cubes with (same-valued) real and lazy data
+ cube_real = Cube(self.data, units="m")
for i_dim, name in enumerate(("y", "x")):
npts = cube_real.shape[i_dim]
coord = DimCoord(np.arange(npts), long_name=name)
cube_real.add_dim_coord(coord, i_dim)
self.cube_real = cube_real
self.cube_lazy = cube_real.copy(data=self.lazydata)
+ self.cube_lazy.units = "kg"
# Test weights and expected result for a y-collapse
self.y_weights = np.array([0.3, 0.5])
self.full_weights_y = np.broadcast_to(
@@ -375,6 +394,7 @@ def test_weighted_fullweights_real_y(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_y
)
+ self.assertEqual(cube_collapsed.units, "m")
def test_weighted_fullweights_lazy_y(self):
# Full-shape weights, lazy data : Check lazy result, same values as real calc.
@@ -385,6 +405,7 @@ def test_weighted_fullweights_lazy_y(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_y
)
+ self.assertEqual(cube_collapsed.units, "kg")
def test_weighted_1dweights_real_y(self):
# 1-D weights, real data : Check same results as full-shape.
@@ -394,6 +415,7 @@ def test_weighted_1dweights_real_y(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_y
)
+ self.assertEqual(cube_collapsed.units, "m")
def test_weighted_1dweights_lazy_y(self):
# 1-D weights, lazy data : Check lazy result, same values as real calc.
@@ -404,6 +426,7 @@ def test_weighted_1dweights_lazy_y(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_y
)
+ self.assertEqual(cube_collapsed.units, "kg")
def test_weighted_fullweights_real_x(self):
# Full weights, real data, ** collapse X ** : as for 'y' case above
@@ -413,6 +436,7 @@ def test_weighted_fullweights_real_x(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_x
)
+ self.assertEqual(cube_collapsed.units, "m")
def test_weighted_fullweights_lazy_x(self):
# Full weights, lazy data, ** collapse X ** : as for 'y' case above
@@ -423,6 +447,7 @@ def test_weighted_fullweights_lazy_x(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_x
)
+ self.assertEqual(cube_collapsed.units, "kg")
def test_weighted_1dweights_real_x(self):
# 1-D weights, real data, ** collapse X ** : as for 'y' case above
@@ -432,6 +457,7 @@ def test_weighted_1dweights_real_x(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_x
)
+ self.assertEqual(cube_collapsed.units, "m")
def test_weighted_1dweights_lazy_x(self):
# 1-D weights, lazy data, ** collapse X ** : as for 'y' case above
@@ -442,6 +468,148 @@ def test_weighted_1dweights_lazy_x(self):
self.assertArrayAlmostEqual(
cube_collapsed.data, self.expected_result_x
)
+ self.assertEqual(cube_collapsed.units, "kg")
+
+ def test_weighted_sum_fullweights_adapt_units_real_y(self):
+ # Check that units are adapted correctly ('m' * '1' = 'm')
+ cube_collapsed = self.cube_real.collapsed(
+ "y", SUM, weights=self.full_weights_y
+ )
+ self.assertEqual(cube_collapsed.units, "m")
+
+ def test_weighted_sum_fullweights_adapt_units_lazy_y(self):
+ # Check that units are adapted correctly ('kg' * '1' = 'kg')
+ cube_collapsed = self.cube_lazy.collapsed(
+ "y", SUM, weights=self.full_weights_y
+ )
+ self.assertEqual(cube_collapsed.units, "kg")
+
+ def test_weighted_sum_1dweights_adapt_units_real_y(self):
+ # Check that units are adapted correctly ('m' * '1' = 'm')
+ # Note: the same test with lazy data fails:
+ # https://github.com/SciTools/iris/issues/5083
+ cube_collapsed = self.cube_real.collapsed(
+ "y", SUM, weights=self.y_weights
+ )
+ self.assertEqual(cube_collapsed.units, "m")
+
+ def test_weighted_sum_with_unknown_units_real_y(self):
+ # Check that units are adapted correctly ('unknown' * '1' = 'unknown')
+ # Note: does not need to be adapted in subclasses since 'unknown'
+ # multiplied by any unit is 'unknown'
+ self.cube_real.units = "unknown"
+ cube_collapsed = self.cube_real.collapsed(
+ "y",
+ SUM,
+ weights=self.full_weights_y,
+ )
+ self.assertEqual(cube_collapsed.units, "unknown")
+
+ def test_weighted_sum_with_unknown_units_lazy_y(self):
+ # Check that units are adapted correctly ('unknown' * '1' = 'unknown')
+ # Note: does not need to be adapted in subclasses since 'unknown'
+ # multiplied by any unit is 'unknown'
+ self.cube_lazy.units = "unknown"
+ cube_collapsed = self.cube_lazy.collapsed(
+ "y",
+ SUM,
+ weights=self.full_weights_y,
+ )
+ self.assertEqual(cube_collapsed.units, "unknown")
+
+
+# Simply redo the tests of Test_collapsed__multidim_weighted_with_arr with
+# other allowed objects for weights
+
+
+class Test_collapsed__multidim_weighted_with_cube(
+ Test_collapsed__multidim_weighted_with_arr
+):
+ def setUp(self):
+ super().setUp()
+
+ self.y_weights_original = self.y_weights
+ self.full_weights_y_original = self.full_weights_y
+ self.x_weights_original = self.x_weights
+ self.full_weights_x_original = self.full_weights_x
+
+ self.y_weights = self.cube_real[:, 0].copy(self.y_weights_original)
+ self.y_weights.units = "m2"
+ self.full_weights_y = self.cube_real.copy(self.full_weights_y_original)
+ self.full_weights_y.units = "m2"
+ self.x_weights = self.cube_real[0, :].copy(self.x_weights_original)
+ self.full_weights_x = self.cube_real.copy(self.full_weights_x_original)
+
+ def test_weighted_sum_fullweights_adapt_units_real_y(self):
+ # Check that units are adapted correctly ('m' * 'm2' = 'm3')
+ cube_collapsed = self.cube_real.collapsed(
+ "y", SUM, weights=self.full_weights_y
+ )
+ self.assertEqual(cube_collapsed.units, "m3")
+
+ def test_weighted_sum_fullweights_adapt_units_lazy_y(self):
+ # Check that units are adapted correctly ('kg' * 'm2' = 'kg m2')
+ cube_collapsed = self.cube_lazy.collapsed(
+ "y", SUM, weights=self.full_weights_y
+ )
+ self.assertEqual(cube_collapsed.units, "kg m2")
+
+ def test_weighted_sum_1dweights_adapt_units_real_y(self):
+ # Check that units are adapted correctly ('m' * 'm2' = 'm3')
+ # Note: the same test with lazy data fails:
+ # https://github.com/SciTools/iris/issues/5083
+ cube_collapsed = self.cube_real.collapsed(
+ "y", SUM, weights=self.y_weights
+ )
+ self.assertEqual(cube_collapsed.units, "m3")
+
+
+class Test_collapsed__multidim_weighted_with_str(
+ Test_collapsed__multidim_weighted_with_cube
+):
+ def setUp(self):
+ super().setUp()
+
+ self.full_weights_y = "full_y"
+ self.full_weights_x = "full_x"
+ self.y_weights = "y"
+ self.x_weights = "1d_x"
+
+ self.dim_metadata_full_y = iris.coords.CellMeasure(
+ self.full_weights_y_original,
+ long_name=self.full_weights_y,
+ units="m2",
+ )
+ self.dim_metadata_full_x = iris.coords.AuxCoord(
+ self.full_weights_x_original,
+ long_name=self.full_weights_x,
+ units="m2",
+ )
+ self.dim_metadata_1d_y = iris.coords.DimCoord(
+ self.y_weights_original, long_name=self.y_weights, units="m2"
+ )
+ self.dim_metadata_1d_x = iris.coords.AncillaryVariable(
+ self.x_weights_original, long_name=self.x_weights, units="m2"
+ )
+
+ for cube in (self.cube_real, self.cube_lazy):
+ cube.add_cell_measure(self.dim_metadata_full_y, (0, 1))
+ cube.add_aux_coord(self.dim_metadata_full_x, (0, 1))
+ cube.remove_coord("y")
+ cube.add_dim_coord(self.dim_metadata_1d_y, 0)
+ cube.add_ancillary_variable(self.dim_metadata_1d_x, 1)
+
+
+class Test_collapsed__multidim_weighted_with_dim_metadata(
+ Test_collapsed__multidim_weighted_with_str
+):
+ def setUp(self):
+ super().setUp()
+
+ self.full_weights_y = self.dim_metadata_full_y
+ self.full_weights_x = self.dim_metadata_full_x
+ self.y_weights = self.dim_metadata_1d_y
+ self.x_weights = self.dim_metadata_1d_x
class Test_collapsed__cellmeasure_ancils(tests.IrisTest):
@@ -501,7 +669,7 @@ def _assert_warn_collapse_without_weight(self, coords, warn):
self.assertIn(mock.call(msg.format(coord)), warn.call_args_list)
def _assert_nowarn_collapse_without_weight(self, coords, warn):
- # Ensure that warning is not rised.
+ # Ensure that warning is not raised.
msg = "Collapsing spatial coordinate {!r} without weighting"
for coord in coords:
self.assertNotIn(mock.call(msg.format(coord)), warn.call_args_list)
@@ -590,7 +758,7 @@ def _assert_warn_cannot_check_contiguity(self, warn):
self.assertIn(mock.call(msg), warn.call_args_list)
def _assert_cube_as_expected(self, cube):
- """Ensure that cube data and coordiantes are as expected."""
+ """Ensure that cube data and coordinates are as expected."""
self.assertArrayEqual(cube.data, np.array(3))
lat = cube.coord("latitude")
@@ -604,16 +772,14 @@ def _assert_cube_as_expected(self, cube):
def test_collapsed_lat_with_3_bounds(self):
"""Collapse latitude with 3 bounds."""
with mock.patch("warnings.warn") as warn:
- collapsed_cube = self.cube.collapsed("latitude", iris.analysis.SUM)
+ collapsed_cube = self.cube.collapsed("latitude", SUM)
self._assert_warn_cannot_check_contiguity(warn)
self._assert_cube_as_expected(collapsed_cube)
def test_collapsed_lon_with_3_bounds(self):
"""Collapse longitude with 3 bounds."""
with mock.patch("warnings.warn") as warn:
- collapsed_cube = self.cube.collapsed(
- "longitude", iris.analysis.SUM
- )
+ collapsed_cube = self.cube.collapsed("longitude", SUM)
self._assert_warn_cannot_check_contiguity(warn)
self._assert_cube_as_expected(collapsed_cube)
@@ -621,7 +787,7 @@ def test_collapsed_lat_lon_with_3_bounds(self):
"""Collapse latitude and longitude with 3 bounds."""
with mock.patch("warnings.warn") as warn:
collapsed_cube = self.cube.collapsed(
- ["latitude", "longitude"], iris.analysis.SUM
+ ["latitude", "longitude"], SUM
)
self._assert_warn_cannot_check_contiguity(warn)
self._assert_cube_as_expected(collapsed_cube)
@@ -741,9 +907,9 @@ def test_different_array_attrs_incompatible(self):
class Test_rolling_window(tests.IrisTest):
def setUp(self):
- self.cube = Cube(np.arange(6))
+ self.cube = Cube(np.arange(6), units="kg")
self.multi_dim_cube = Cube(np.arange(36).reshape(6, 6))
- val_coord = DimCoord([0, 1, 2, 3, 4, 5], long_name="val")
+ val_coord = DimCoord([0, 1, 2, 3, 4, 5], long_name="val", units="s")
month_coord = AuxCoord(
["jan", "feb", "mar", "apr", "may", "jun"], long_name="month"
)
@@ -770,6 +936,7 @@ def test_string_coord(self):
np.array([1, 2, 3, 4]),
bounds=np.array([[0, 2], [1, 3], [2, 4], [3, 5]]),
long_name="val",
+ units="s",
)
month_coord = AuxCoord(
np.array(
@@ -818,6 +985,30 @@ def test_ancillary_variables_and_cell_measures_removed(self):
self.assertEqual(res_cube.ancillary_variables(), [])
self.assertEqual(res_cube.cell_measures(), [])
+ def test_weights_arr(self):
+ weights = [0, 0, 1, 0, 2]
+ res_cube = self.cube.rolling_window("val", SUM, 5, weights=weights)
+ np.testing.assert_array_equal(res_cube.data, [10, 13])
+ self.assertEqual(res_cube.units, "kg")
+
+ def test_weights_cube(self):
+ weights = Cube([0, 0, 1, 0, 2], units="m2")
+ res_cube = self.cube.rolling_window("val", SUM, 5, weights=weights)
+ np.testing.assert_array_equal(res_cube.data, [10, 13])
+ self.assertEqual(res_cube.units, "kg m2")
+
+ def test_weights_str(self):
+ weights = "val"
+ res_cube = self.cube.rolling_window("val", SUM, 6, weights=weights)
+ np.testing.assert_array_equal(res_cube.data, [55])
+ self.assertEqual(res_cube.units, "kg s")
+
+ def test_weights_dim_coord(self):
+ weights = self.cube.coord("val")
+ res_cube = self.cube.rolling_window("val", SUM, 6, weights=weights)
+ np.testing.assert_array_equal(res_cube.data, [55])
+ self.assertEqual(res_cube.units, "kg s")
+
class Test_slices_dim_order(tests.IrisTest):
"""
@@ -905,7 +1096,7 @@ def setUp(self):
len(self.cube.coord("model_level_number").points)
)
self.exp_iter_2d = np.ndindex(6, 70, 1, 1)
- # Define maximum number of interations for particularly long
+ # Define maximum number of interactions for particularly long
# (and so time-consuming) iterators.
self.long_iterator_max = 5
@@ -1962,6 +2153,7 @@ def _assert_lists_equal(self, items_a, items_b):
a different order.
"""
+
# Compare (and thus sort) by their *common* metadata.
def sortkey(item):
return BaseMetadata.from_metadata(item.metadata)
diff --git a/lib/iris/tests/unit/experimental/ugrid/cf/test_CFUGridReader.py b/lib/iris/tests/unit/experimental/ugrid/cf/test_CFUGridReader.py
index e44aee730a..d9de814b05 100644
--- a/lib/iris/tests/unit/experimental/ugrid/cf/test_CFUGridReader.py
+++ b/lib/iris/tests/unit/experimental/ugrid/cf/test_CFUGridReader.py
@@ -94,7 +94,10 @@ def setUp(self):
# Restrict the CFUGridReader functionality to only performing
# translations and building first level cf-groups for variables.
self.patch("iris.experimental.ugrid.cf.CFUGridReader._reset")
- self.patch("netCDF4.Dataset", return_value=self.dataset)
+ self.patch(
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
+ )
cf_reader = CFUGridReader("dummy")
self.cf_group = cf_reader.cf_group
diff --git a/lib/iris/tests/unit/experimental/ugrid/mesh/test_MeshCoord.py b/lib/iris/tests/unit/experimental/ugrid/mesh/test_MeshCoord.py
index 538cecdc7d..03e2793fd9 100644
--- a/lib/iris/tests/unit/experimental/ugrid/mesh/test_MeshCoord.py
+++ b/lib/iris/tests/unit/experimental/ugrid/mesh/test_MeshCoord.py
@@ -235,9 +235,7 @@ class Test__getitem__(tests.IrisTest):
def test_slice_wholeslice_1tuple(self):
# The only slicing case that we support, to enable cube slicing.
meshcoord = sample_meshcoord()
- meshcoord2 = meshcoord[
- :,
- ]
+ meshcoord2 = meshcoord[:,]
self.assertIsNot(meshcoord2, meshcoord)
self.assertEqual(meshcoord2, meshcoord)
# In this case, we should *NOT* copy the linked Mesh object.
diff --git a/lib/iris/tests/unit/fileformats/cf/test_CFReader.py b/lib/iris/tests/unit/fileformats/cf/test_CFReader.py
index dee28e98cc..9e5cf9b7a5 100644
--- a/lib/iris/tests/unit/fileformats/cf/test_CFReader.py
+++ b/lib/iris/tests/unit/fileformats/cf/test_CFReader.py
@@ -70,7 +70,10 @@ def setUp(self):
)
def test_create_global_attributes(self):
- with mock.patch("netCDF4.Dataset", return_value=self.dataset):
+ with mock.patch(
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
+ ):
global_attrs = CFReader("dummy").cf_group.global_attributes
self.assertEqual(
global_attrs["dimensions"], "something something_else"
@@ -145,7 +148,10 @@ def setUp(self):
self.addCleanup(reset_patch.stop)
def test_create_formula_terms(self):
- with mock.patch("netCDF4.Dataset", return_value=self.dataset):
+ with mock.patch(
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
+ ):
cf_group = CFReader("dummy").cf_group
self.assertEqual(len(cf_group), len(self.variables))
# Check there is a singular data variable.
@@ -247,7 +253,10 @@ def setUp(self):
self.addCleanup(patcher.stop)
def test_associate_formula_terms_with_data_variable(self):
- with mock.patch("netCDF4.Dataset", return_value=self.dataset):
+ with mock.patch(
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
+ ):
cf_group = CFReader("dummy").cf_group
self.assertEqual(len(cf_group), len(self.variables))
# Check the cf-group associated with the data variable.
@@ -296,7 +305,10 @@ def test_associate_formula_terms_with_data_variable(self):
)
def test_promote_reference(self):
- with mock.patch("netCDF4.Dataset", return_value=self.dataset):
+ with mock.patch(
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
+ ):
cf_group = CFReader("dummy").cf_group
self.assertEqual(len(cf_group), len(self.variables))
# Check the number of data variables.
@@ -316,7 +328,8 @@ def test_promote_reference(self):
def test_formula_terms_ignore(self):
self.orography.dimensions = ["lat", "wibble"]
with mock.patch(
- "netCDF4.Dataset", return_value=self.dataset
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
), mock.patch("warnings.warn") as warn:
cf_group = CFReader("dummy").cf_group
group = cf_group.promoted
@@ -327,7 +340,8 @@ def test_formula_terms_ignore(self):
def test_auxiliary_ignore(self):
self.x.dimensions = ["lat", "wibble"]
with mock.patch(
- "netCDF4.Dataset", return_value=self.dataset
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
), mock.patch("warnings.warn") as warn:
cf_group = CFReader("dummy").cf_group
promoted = ["x", "orography"]
@@ -342,7 +356,8 @@ def test_promoted_auxiliary_ignore(self):
self.variables["wibble"] = self.wibble
self.orography.coordinates = "wibble"
with mock.patch(
- "netCDF4.Dataset", return_value=self.dataset
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper",
+ return_value=self.dataset,
), mock.patch("warnings.warn") as warn:
cf_group = CFReader("dummy").cf_group.promoted
promoted = ["wibble", "orography"]
diff --git a/lib/iris/tests/unit/fileformats/nc_load_rules/actions/__init__.py b/lib/iris/tests/unit/fileformats/nc_load_rules/actions/__init__.py
index 0cc3d09426..399a987f11 100644
--- a/lib/iris/tests/unit/fileformats/nc_load_rules/actions/__init__.py
+++ b/lib/iris/tests/unit/fileformats/nc_load_rules/actions/__init__.py
@@ -80,42 +80,44 @@ def load_cube_from_cdl(self, cdl_string, cdl_path, nc_path):
# Simulate the inner part of the file reading process.
cf = CFReader(nc_path)
- # Grab a data variable : FOR NOW always grab the 'phenom' variable.
- cf_var = cf.cf_group.data_variables["phenom"]
-
- engine = iris.fileformats.netcdf.loader._actions_engine()
-
- # If debug enabled, switch on the activation summary debug output.
- # Use 'patch' so it is restored after the test.
- self.patch("iris.fileformats.netcdf.loader.DEBUG", self.debug)
-
- with warnings.catch_warnings():
- warnings.filterwarnings(
- "ignore",
- message="Ignoring a datum in netCDF load for consistency with existing "
- "behaviour. In a future version of Iris, this datum will be "
- "applied. To apply the datum when loading, use the "
- "iris.FUTURE.datum_support flag.",
- category=FutureWarning,
- )
- # Call the main translation function to load a single cube.
- # _load_cube establishes per-cube facts, activates rules and
- # produces an actual cube.
- cube = _load_cube(engine, cf, cf_var, nc_path)
-
- # Also Record, on the cubes, which hybrid coord elements were identified
- # by the rules operation.
- # Unlike the other translations, _load_cube does *not* convert this
- # information into actual cube elements. That is instead done by
- # `iris.fileformats.netcdf._load_aux_factory`.
- # For rules testing, it is anyway more convenient to deal with the raw
- # data, as each factory type has different validity requirements to
- # build it, and none of that is relevant to the rules operation.
- cube._formula_type_name = engine.requires.get("formula_type")
- cube._formula_terms_byname = engine.requires.get("formula_terms")
-
- # Always returns a single cube.
- return cube
+
+ with cf:
+ # Grab a data variable : FOR NOW always grab the 'phenom' variable.
+ cf_var = cf.cf_group.data_variables["phenom"]
+
+ engine = iris.fileformats.netcdf.loader._actions_engine()
+
+ # If debug enabled, switch on the activation summary debug output.
+ # Use 'patch' so it is restored after the test.
+ self.patch("iris.fileformats.netcdf.loader.DEBUG", self.debug)
+
+ with warnings.catch_warnings():
+ warnings.filterwarnings(
+ "ignore",
+ message="Ignoring a datum in netCDF load for consistency with existing "
+ "behaviour. In a future version of Iris, this datum will be "
+ "applied. To apply the datum when loading, use the "
+ "iris.FUTURE.datum_support flag.",
+ category=FutureWarning,
+ )
+ # Call the main translation function to load a single cube.
+ # _load_cube establishes per-cube facts, activates rules and
+ # produces an actual cube.
+ cube = _load_cube(engine, cf, cf_var, nc_path)
+
+ # Also Record, on the cubes, which hybrid coord elements were identified
+ # by the rules operation.
+ # Unlike the other translations, _load_cube does *not* convert this
+ # information into actual cube elements. That is instead done by
+ # `iris.fileformats.netcdf._load_aux_factory`.
+ # For rules testing, it is anyway more convenient to deal with the raw
+ # data, as each factory type has different validity requirements to
+ # build it, and none of that is relevant to the rules operation.
+ cube._formula_type_name = engine.requires.get("formula_type")
+ cube._formula_terms_byname = engine.requires.get("formula_terms")
+
+ # Always returns a single cube.
+ return cube
def run_testcase(self, warning_regex=None, **testcase_kwargs):
"""
diff --git a/lib/iris/tests/unit/fileformats/nc_load_rules/actions/test__miscellaneous.py b/lib/iris/tests/unit/fileformats/nc_load_rules/actions/test__miscellaneous.py
index a8e44747dd..ffe00c8c19 100644
--- a/lib/iris/tests/unit/fileformats/nc_load_rules/actions/test__miscellaneous.py
+++ b/lib/iris/tests/unit/fileformats/nc_load_rules/actions/test__miscellaneous.py
@@ -127,7 +127,6 @@ def _make_testcase_cdl(
include_cellmeasure=False,
include_ancil=False,
):
-
phenom_extra_attrs_string = ""
extra_vars_string = ""
diff --git a/lib/iris/tests/unit/fileformats/nc_load_rules/helpers/test_build_dimension_coordinate.py b/lib/iris/tests/unit/fileformats/nc_load_rules/helpers/test_build_dimension_coordinate.py
index b485937cb1..bc13975441 100644
--- a/lib/iris/tests/unit/fileformats/nc_load_rules/helpers/test_build_dimension_coordinate.py
+++ b/lib/iris/tests/unit/fileformats/nc_load_rules/helpers/test_build_dimension_coordinate.py
@@ -229,7 +229,9 @@ def test_aux_coord_construction(self):
warning_patch = mock.patch("warnings.warn")
# Asserts must lie within context manager because of deferred loading.
- with warning_patch, self.deferred_load_patch, self.get_cf_bounds_var_patch:
+ with (
+ warning_patch
+ ), self.deferred_load_patch, self.get_cf_bounds_var_patch:
build_dimension_coordinate(self.engine, self.cf_coord_var)
# Test that expected coord is built and added to cube.
diff --git a/lib/iris/tests/unit/fileformats/netcdf/test_Saver.py b/lib/iris/tests/unit/fileformats/netcdf/test_Saver.py
index 174a46fdb7..93a1537ea4 100644
--- a/lib/iris/tests/unit/fileformats/netcdf/test_Saver.py
+++ b/lib/iris/tests/unit/fileformats/netcdf/test_Saver.py
@@ -13,7 +13,6 @@
from contextlib import contextmanager
from unittest import mock
-import netCDF4 as nc
import numpy as np
from numpy import ma
@@ -32,7 +31,7 @@
)
from iris.coords import AuxCoord, DimCoord
from iris.cube import Cube
-from iris.fileformats.netcdf import Saver
+from iris.fileformats.netcdf import Saver, _thread_safe_nc
import iris.tests.stock as stock
@@ -203,12 +202,12 @@ def test_big_endian(self):
def test_zlib(self):
cube = self._simple_cube(">f4")
- api = self.patch("iris.fileformats.netcdf.saver.netCDF4")
+ api = self.patch("iris.fileformats.netcdf.saver._thread_safe_nc")
# Define mocked default fill values to prevent deprecation warning (#4374).
api.default_fillvals = collections.defaultdict(lambda: -99.0)
with Saver("/dummy/path", "NETCDF4") as saver:
saver.write(cube, zlib=True)
- dataset = api.Dataset.return_value
+ dataset = api.DatasetWrapper.return_value
create_var_call = mock.call(
"air_pressure_anomaly",
np.dtype("float32"),
@@ -249,7 +248,7 @@ def test_default_unlimited_dimensions(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube)
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
self.assertFalse(ds.dimensions["dim0"].isunlimited())
self.assertFalse(ds.dimensions["dim1"].isunlimited())
ds.close()
@@ -259,7 +258,7 @@ def test_no_unlimited_dimensions(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=None)
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
for dim in ds.dimensions.values():
self.assertFalse(dim.isunlimited())
ds.close()
@@ -281,7 +280,7 @@ def test_custom_unlimited_dimensions(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=unlimited_dimensions)
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
for dim in unlimited_dimensions:
self.assertTrue(ds.dimensions[dim].isunlimited())
ds.close()
@@ -290,7 +289,7 @@ def test_custom_unlimited_dimensions(self):
coords = [cube.coord(dim) for dim in unlimited_dimensions]
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=coords)
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
for dim in unlimited_dimensions:
self.assertTrue(ds.dimensions[dim].isunlimited())
ds.close()
@@ -301,7 +300,7 @@ def test_reserved_attributes(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube)
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
res = ds.getncattr("dimensions")
ds.close()
self.assertEqual(res, "something something_else")
@@ -323,7 +322,7 @@ def test_dimensional_to_scalar(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube)
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
# Confirm that the only dimension is the one denoting the number
# of bounds - have successfully saved the 2D bounds array into 1D.
self.assertEqual(["bnds"], list(ds.dimensions.keys()))
@@ -363,7 +362,7 @@ def _check_bounds_setting(self, climatological=False):
saver._ensure_valid_dtype.return_value = mock.Mock(
shape=coord.bounds.shape, dtype=coord.bounds.dtype
)
- var = mock.MagicMock(spec=nc.Variable)
+ var = mock.MagicMock(spec=_thread_safe_nc.VariableWrapper)
# Make the main call.
Saver._create_cf_bounds(saver, coord, var, "time")
@@ -404,7 +403,7 @@ def test_valid_range_saved(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=[])
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
self.assertArrayEqual(ds.valid_range, vrange)
ds.close()
@@ -416,7 +415,7 @@ def test_valid_min_saved(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=[])
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
self.assertArrayEqual(ds.valid_min, 1)
ds.close()
@@ -428,7 +427,7 @@ def test_valid_max_saved(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=[])
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
self.assertArrayEqual(ds.valid_max, 2)
ds.close()
@@ -448,7 +447,7 @@ def test_valid_range_saved(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=[])
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
self.assertArrayEqual(
ds.variables["longitude"].valid_range, vrange
)
@@ -462,7 +461,7 @@ def test_valid_min_saved(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=[])
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
self.assertArrayEqual(ds.variables["longitude"].valid_min, 1)
ds.close()
@@ -474,7 +473,7 @@ def test_valid_max_saved(self):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, unlimited_dimensions=[])
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
self.assertArrayEqual(ds.variables["longitude"].valid_max, 2)
ds.close()
@@ -506,7 +505,7 @@ def _netCDF_var(self, cube, **kwargs):
with self.temp_filename(".nc") as nc_path:
with Saver(nc_path, "NETCDF4") as saver:
saver.write(cube, **kwargs)
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
(var,) = [
var
for var in ds.variables.values()
@@ -572,7 +571,7 @@ def test_contains_default_fill_value(self):
# Test that a warning is raised if the data contains the default fill
# value if no fill_value argument is supplied.
cube = self._make_cube(">f4")
- cube.data[0, 0] = nc.default_fillvals["f4"]
+ cube.data[0, 0] = _thread_safe_nc.default_fillvals["f4"]
with self.assertWarnsRegex(
UserWarning,
"contains unmasked data points equal to the fill-value",
@@ -647,7 +646,9 @@ def setUp(self):
self.container = mock.Mock(name="container", attributes={})
self.data_dtype = np.dtype("int32")
- patch = mock.patch("netCDF4.Dataset")
+ patch = mock.patch(
+ "iris.fileformats.netcdf._thread_safe_nc.DatasetWrapper"
+ )
_ = patch.start()
self.addCleanup(patch.stop)
@@ -1050,38 +1051,5 @@ def test_geo_cs(self):
self._test(coord_system, expected)
-class Test__create_cf_cell_measure_variable(tests.IrisTest):
- # Saving of masked data is disallowed.
-
- # Attribute is substituted in test_Saver__lazy.
- array_lib = np
-
- def setUp(self):
- self.cube = stock.lat_lon_cube()
- self.names_map = ["latitude", "longitude"]
- masked_array = self.array_lib.ma.masked_array(
- [0, 1, 2], mask=[True, False, True]
- )
- self.cm = iris.coords.CellMeasure(masked_array, var_name="cell_area")
- self.cube.add_cell_measure(self.cm, data_dims=0)
- self.exp_emsg = "Cell measures with missing data are not supported."
-
- def test_masked_data__insitu(self):
- # Test that the error is raised in the right place.
- with self.temp_filename(".nc") as nc_path:
- saver = Saver(nc_path, "NETCDF4")
- with self.assertRaisesRegex(ValueError, self.exp_emsg):
- saver._create_generic_cf_array_var(
- self.cube, self.names_map, self.cm
- )
-
- def test_masked_data__save_pipeline(self):
- # Test that the right error is raised by the saver pipeline.
- with self.temp_filename(".nc") as nc_path:
- with Saver(nc_path, "NETCDF4") as saver:
- with self.assertRaisesRegex(ValueError, self.exp_emsg):
- saver.write(self.cube)
-
-
if __name__ == "__main__":
tests.main()
diff --git a/lib/iris/tests/unit/fileformats/netcdf/test_Saver__lazy.py b/lib/iris/tests/unit/fileformats/netcdf/test_Saver__lazy.py
index eab09b9e4f..53e1f9a652 100644
--- a/lib/iris/tests/unit/fileformats/netcdf/test_Saver__lazy.py
+++ b/lib/iris/tests/unit/fileformats/netcdf/test_Saver__lazy.py
@@ -82,12 +82,6 @@ class Test_check_attribute_compliance__exception_handling(
pass
-class Test__create_cf_cell_measure_variable(
- LazyMixin, test_Saver.Test__create_cf_cell_measure_variable
-):
- pass
-
-
class TestStreamed(tests.IrisTest):
def setUp(self):
self.cube = stock.simple_2d()
diff --git a/lib/iris/tests/unit/fileformats/netcdf/test_Saver__ugrid.py b/lib/iris/tests/unit/fileformats/netcdf/test_Saver__ugrid.py
index 575c852ece..323b498d9c 100644
--- a/lib/iris/tests/unit/fileformats/netcdf/test_Saver__ugrid.py
+++ b/lib/iris/tests/unit/fileformats/netcdf/test_Saver__ugrid.py
@@ -18,7 +18,6 @@
import shutil
import tempfile
-import netCDF4 as nc
import numpy as np
from iris import save
@@ -26,6 +25,7 @@
from iris.cube import Cube, CubeList
from iris.experimental.ugrid.mesh import Connectivity, Mesh
from iris.experimental.ugrid.save import save_mesh
+from iris.fileformats.netcdf import _thread_safe_nc
from iris.tests.stock import realistic_4d
XY_LOCS = ("x", "y")
@@ -259,7 +259,7 @@ def scan_dataset(filepath):
variable's dims.
"""
- ds = nc.Dataset(filepath)
+ ds = _thread_safe_nc.DatasetWrapper(filepath)
# dims dict is {name: len}
dimsdict = {name: dim.size for name, dim in ds.dimensions.items()}
# vars dict is {name: {attr:val}}
@@ -824,7 +824,7 @@ def test_nonuniform_connectivity(self):
self.assertNotIn("_FillValue", fn_props)
# For what it's worth, *also* check the actual data array in the file
- ds = nc.Dataset(tempfile_path)
+ ds = _thread_safe_nc.DatasetWrapper(tempfile_path)
conn_var = ds.variables[ff_conn_name]
data = conn_var[:]
ds.close()
@@ -1082,7 +1082,6 @@ def test_mesh_dim_names(self):
("dim invalid-name &%!", "dim_invalid_name____"),
]
for given_name, expected_name in dim_names_tests:
-
mesh = make_mesh(mesh_kwargs={"face_dimension": given_name})
filepath = self.check_save_mesh(mesh)
diff --git a/lib/iris/tests/unit/fileformats/netcdf/test_save.py b/lib/iris/tests/unit/fileformats/netcdf/test_save.py
index 030edbfce2..b274a8be0d 100644
--- a/lib/iris/tests/unit/fileformats/netcdf/test_save.py
+++ b/lib/iris/tests/unit/fileformats/netcdf/test_save.py
@@ -14,14 +14,17 @@
from tempfile import mkdtemp
from unittest import mock
-import netCDF4 as nc
import numpy as np
import iris
from iris.coords import AuxCoord, DimCoord
from iris.cube import Cube, CubeList
from iris.experimental.ugrid import PARSE_UGRID_ON_LOAD
-from iris.fileformats.netcdf import CF_CONVENTIONS_VERSION, save
+from iris.fileformats.netcdf import (
+ CF_CONVENTIONS_VERSION,
+ _thread_safe_nc,
+ save,
+)
from iris.tests.stock import lat_lon_cube
from iris.tests.stock.mesh import sample_mesh_cube
@@ -38,7 +41,7 @@ def test_custom_conventions__ignored(self):
# CF convention.
with self.temp_filename(".nc") as nc_path:
save(self.cube, nc_path, "NETCDF4")
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
res = ds.getncattr("Conventions")
ds.close()
self.assertEqual(res, CF_CONVENTIONS_VERSION)
@@ -49,7 +52,7 @@ def test_custom_conventions__allowed(self):
with mock.patch.object(self.options, "conventions_override", True):
with self.temp_filename(".nc") as nc_path:
save(self.cube, nc_path, "NETCDF4")
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
res = ds.getncattr("Conventions")
ds.close()
self.assertEqual(res, self.custom_conventions)
@@ -61,7 +64,7 @@ def test_custom_conventions__allowed__missing(self):
with mock.patch.object(self.options, "conventions_override", True):
with self.temp_filename(".nc") as nc_path:
save(self.cube, nc_path, "NETCDF4")
- ds = nc.Dataset(nc_path)
+ ds = _thread_safe_nc.DatasetWrapper(nc_path)
res = ds.getncattr("Conventions")
ds.close()
self.assertEqual(res, CF_CONVENTIONS_VERSION)
@@ -76,7 +79,7 @@ def test_attributes_arrays(self):
with self.temp_filename("foo.nc") as nc_out:
save([c1, c2], nc_out)
- ds = nc.Dataset(nc_out)
+ ds = _thread_safe_nc.DatasetWrapper(nc_out)
res = ds.getncattr("bar")
ds.close()
self.assertArrayEqual(res, np.arange(2))
@@ -92,7 +95,7 @@ def test_no_special_attribute_clash(self):
with self.temp_filename("foo.nc") as nc_out:
save([c1, c2], nc_out)
- ds = nc.Dataset(nc_out)
+ ds = _thread_safe_nc.DatasetWrapper(nc_out)
res = ds.variables["test"].getncattr("name")
res_1 = ds.variables["test_1"].getncattr("name")
ds.close()
@@ -105,7 +108,7 @@ def test_no_unlimited_dims(self):
cube = lat_lon_cube()
with self.temp_filename("foo.nc") as nc_out:
save(cube, nc_out)
- ds = nc.Dataset(nc_out)
+ ds = _thread_safe_nc.DatasetWrapper(nc_out)
self.assertFalse(ds.dimensions["latitude"].isunlimited())
def test_unlimited_dim_latitude(self):
@@ -113,7 +116,7 @@ def test_unlimited_dim_latitude(self):
unlim_dim_name = "latitude"
with self.temp_filename("foo.nc") as nc_out:
save(cube, nc_out, unlimited_dimensions=[unlim_dim_name])
- ds = nc.Dataset(nc_out)
+ ds = _thread_safe_nc.DatasetWrapper(nc_out)
self.assertTrue(ds.dimensions[unlim_dim_name].isunlimited())
diff --git a/lib/iris/tests/unit/fileformats/pp/test_PPField.py b/lib/iris/tests/unit/fileformats/pp/test_PPField.py
index 5e2bbcaa2c..316894ded1 100644
--- a/lib/iris/tests/unit/fileformats/pp/test_PPField.py
+++ b/lib/iris/tests/unit/fileformats/pp/test_PPField.py
@@ -44,7 +44,6 @@
class DummyPPField(PPField):
-
HEADER_DEFN = DUMMY_HEADER
HEADER_DICT = dict(DUMMY_HEADER)
diff --git a/lib/iris/tests/unit/fileformats/pp_load_rules/test__all_other_rules.py b/lib/iris/tests/unit/fileformats/pp_load_rules/test__all_other_rules.py
index 62eb7ff019..e194e240c6 100644
--- a/lib/iris/tests/unit/fileformats/pp_load_rules/test__all_other_rules.py
+++ b/lib/iris/tests/unit/fileformats/pp_load_rules/test__all_other_rules.py
@@ -211,7 +211,6 @@ def test_lbcode3x23(self):
class TestLBTIMx2x_ZeroYears(TestField):
-
_spec = [
"lbtim",
"lbcode",
diff --git a/lib/iris/tests/unit/lazy_data/test_as_lazy_data.py b/lib/iris/tests/unit/lazy_data/test_as_lazy_data.py
index 5aeebd6045..5f9dece153 100644
--- a/lib/iris/tests/unit/lazy_data/test_as_lazy_data.py
+++ b/lib/iris/tests/unit/lazy_data/test_as_lazy_data.py
@@ -68,7 +68,7 @@ def test_chunk_size_limiting(self):
((11, 2, 1011, 1022), (5, 2, 1011, 1022)),
]
err_fmt = "Result of optimising chunks {} was {}, expected {}"
- for (shape, expected) in given_shapes_and_resulting_chunks:
+ for shape, expected in given_shapes_and_resulting_chunks:
chunks = _optimum_chunksize(
shape, shape, limit=self.FIXED_CHUNKSIZE_LIMIT
)
@@ -86,7 +86,7 @@ def test_chunk_size_expanding(self):
((3, 300, 200), (117, 300, 1000), (39, 300, 1000)),
]
err_fmt = "Result of optimising shape={};chunks={} was {}, expected {}"
- for (shape, fullshape, expected) in given_shapes_and_resulting_chunks:
+ for shape, fullshape, expected in given_shapes_and_resulting_chunks:
chunks = _optimum_chunksize(
chunks=shape, shape=fullshape, limit=self.FIXED_CHUNKSIZE_LIMIT
)
diff --git a/lib/iris/tests/unit/merge/test_ProtoCube.py b/lib/iris/tests/unit/merge/test_ProtoCube.py
index 625290ad24..0fca726b28 100644
--- a/lib/iris/tests/unit/merge/test_ProtoCube.py
+++ b/lib/iris/tests/unit/merge/test_ProtoCube.py
@@ -289,7 +289,6 @@ def test_noise(self):
class Test_register__CoordSig_general(_MergeTest, tests.IrisTest):
-
_mergetest_type = "coord"
def setUp(self):
@@ -444,7 +443,6 @@ def test_coord_system(self):
class Test_register__CoordSig_scalar(_MergeTest_coordprops, tests.IrisTest):
-
_mergetest_type = "aux_coords (scalar)"
def setUp(self):
@@ -486,7 +484,6 @@ def test_dims(self):
class Test_register__CoordSig_dim(_MergeTest_coordprops_vect, tests.IrisTest):
-
_mergetest_type = "dim_coords"
_coord_typename = "dim_coord"
@@ -515,7 +512,6 @@ def test_circular(self):
class Test_register__CoordSig_aux(_MergeTest_coordprops_vect, tests.IrisTest):
-
_mergetest_type = "aux_coords (non-scalar)"
_coord_typename = "aux_coord"
diff --git a/lib/iris/util.py b/lib/iris/util.py
index 3d82ea68c5..9e0db9e66e 100644
--- a/lib/iris/util.py
+++ b/lib/iris/util.py
@@ -735,7 +735,6 @@ def _build_full_slice_given_keys(keys, ndim):
for i, key in enumerate(keys):
if key is Ellipsis:
-
# replace any subsequent Ellipsis objects in keys with
# slice(None, None) as per Numpy
keys = keys[:i] + tuple(
@@ -1815,8 +1814,9 @@ def _mask_array(array, points_to_mask, in_place=False):
If array is lazy then in_place is ignored: _math_op_common will use the
returned value regardless of in_place, so we do not need to implement it
- here. If in_place is True then array must be a np.ma.MaskedArray or dask
- array (must be a dask array if points_to_mask is lazy).
+ here. If in_place is True then array must be a
+ :class:`numpy.ma.MaskedArray` or :class:`dask.array.Array`
+ (must be a dask array if points_to_mask is lazy).
"""
# Decide which array library to use.
@@ -1978,7 +1978,7 @@ def is_masked(array):
Parameters
----------
- array : :class:`numpy.Array` or `dask.array.Array`
+ array : :class:`numpy.Array` or :class:`dask.array.Array`
The array to be checked for masks.
Returns
diff --git a/noxfile.py b/noxfile.py
index 8aabf862fb..c7b0a0e05b 100755
--- a/noxfile.py
+++ b/noxfile.py
@@ -176,6 +176,8 @@ def tests(session: nox.sessions.Session):
"""
Perform iris system, integration and unit tests.
+ Coverage testing is enabled if the "--coverage" or "-c" flag is used.
+
Parameters
----------
session: object
@@ -185,13 +187,15 @@ def tests(session: nox.sessions.Session):
prepare_venv(session)
session.install("--no-deps", "--editable", ".")
session.env.update(ENV)
- session.run(
+ run_args = [
"python",
"-m",
"iris.tests.runner",
"--default-tests",
- "--system-tests",
- )
+ ]
+ if "-c" in session.posargs or "--coverage" in session.posargs:
+ run_args.append("--coverage")
+ session.run(*run_args)
@nox.session(python=_PY_VERSION_DOCSBUILD, venv_backend="conda")
diff --git a/pyproject.toml b/pyproject.toml
index bdb8a431e5..b44187191b 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -46,3 +46,19 @@ verbose = "False"
[tool.pytest.ini_options]
addopts = "-ra"
testpaths = "lib/iris"
+
+[tool.coverage.run]
+branch = true
+source = [
+ "lib/iris",
+]
+omit = [
+ "lib/iris/tests/*",
+ "lib/iris/etc/*",
+]
+
+[tool.coverage.report]
+exclude_lines = [
+ "pragma: no cover",
+ "if __name__ == .__main__.:"
+]
diff --git a/requirements/ci/nox.lock/py310-linux-64.lock b/requirements/ci/nox.lock/py310-linux-64.lock
index 81a1661c1e..6a334b811c 100644
--- a/requirements/ci/nox.lock/py310-linux-64.lock
+++ b/requirements/ci/nox.lock/py310-linux-64.lock
@@ -1,6 +1,6 @@
# Generated by conda-lock.
# platform: linux-64
-# input_hash: 65e8c3d4ababc804f8d3715a14ce94c3f564a37860525f35ea1aed69efd67be8
+# input_hash: f8af5f4aafcb766f463a1a897d3dab9e04f05f1494bced5931d78175ca0c66df
@EXPLICIT
https://conda.anaconda.org/conda-forge/linux-64/_libgcc_mutex-0.1-conda_forge.tar.bz2#d7c89558ba9fa0495403155b64376d81
https://conda.anaconda.org/conda-forge/linux-64/ca-certificates-2022.12.7-ha878542_0.conda#ff9f73d45c4a07d6f424495288a26080
@@ -8,7 +8,7 @@ https://conda.anaconda.org/conda-forge/noarch/font-ttf-dejavu-sans-mono-2.37-hab
https://conda.anaconda.org/conda-forge/noarch/font-ttf-inconsolata-3.000-h77eed37_0.tar.bz2#34893075a5c9e55cdafac56607368fc6
https://conda.anaconda.org/conda-forge/noarch/font-ttf-source-code-pro-2.038-h77eed37_0.tar.bz2#4d59c254e01d9cde7957100457e2d5fb
https://conda.anaconda.org/conda-forge/noarch/font-ttf-ubuntu-0.83-hab24e00_0.tar.bz2#19410c3df09dfb12d1206132a1d357c5
-https://conda.anaconda.org/conda-forge/linux-64/ld_impl_linux-64-2.39-hcc3a1bd_1.conda#737be0d34c22d24432049ab7a3214de4
+https://conda.anaconda.org/conda-forge/linux-64/ld_impl_linux-64-2.40-h41732ed_0.conda#7aca3059a1729aa76c597603f10b0dd3
https://conda.anaconda.org/conda-forge/linux-64/libgfortran5-12.2.0-h337968e_19.tar.bz2#164b4b1acaedc47ee7e658ae6b308ca3
https://conda.anaconda.org/conda-forge/linux-64/libstdcxx-ng-12.2.0-h46fd767_19.tar.bz2#1030b1f38c129f2634eae026f704fe60
https://conda.anaconda.org/conda-forge/linux-64/mpi-1.0-mpich.tar.bz2#c1fcff3417b5a22bbc4cf6e8c23648cf
@@ -33,13 +33,14 @@ https://conda.anaconda.org/conda-forge/linux-64/giflib-5.2.1-h36c2ea0_2.tar.bz2#
https://conda.anaconda.org/conda-forge/linux-64/graphite2-1.3.13-h58526e2_1001.tar.bz2#8c54672728e8ec6aa6db90cf2806d220
https://conda.anaconda.org/conda-forge/linux-64/gstreamer-orc-0.4.33-h166bdaf_0.tar.bz2#879c93426c9d0b84a9de4513fbce5f4f
https://conda.anaconda.org/conda-forge/linux-64/icu-70.1-h27087fc_0.tar.bz2#87473a15119779e021c314249d4b4aed
-https://conda.anaconda.org/conda-forge/linux-64/jpeg-9e-h166bdaf_2.tar.bz2#ee8b844357a0946870901c7c6f418268
+https://conda.anaconda.org/conda-forge/linux-64/jpeg-9e-h0b41bf4_3.conda#c7a069243e1fbe9a556ed2ec030e6407
https://conda.anaconda.org/conda-forge/linux-64/keyutils-1.6.1-h166bdaf_0.tar.bz2#30186d27e2c9fa62b45fb1476b7200e3
https://conda.anaconda.org/conda-forge/linux-64/lame-3.100-h166bdaf_1003.tar.bz2#a8832b479f93521a9e7b5b743803be51
https://conda.anaconda.org/conda-forge/linux-64/lerc-4.0.0-h27087fc_0.tar.bz2#76bbff344f0134279f225174e9064c8f
+https://conda.anaconda.org/conda-forge/linux-64/libaec-1.0.6-hcb278e6_1.conda#0f683578378cddb223e7fd24f785ab2a
https://conda.anaconda.org/conda-forge/linux-64/libbrotlicommon-1.0.9-h166bdaf_8.tar.bz2#9194c9bf9428035a05352d031462eae4
https://conda.anaconda.org/conda-forge/linux-64/libdb-6.2.32-h9c3ff4c_0.tar.bz2#3f3258d8f841fbac63b36b75bdac1afd
-https://conda.anaconda.org/conda-forge/linux-64/libdeflate-1.14-h166bdaf_0.tar.bz2#fc84a0446e4e4fb882e78d786cfb9734
+https://conda.anaconda.org/conda-forge/linux-64/libdeflate-1.17-h0b41bf4_0.conda#5cc781fd91968b11a8a7fdbee0982676
https://conda.anaconda.org/conda-forge/linux-64/libev-4.33-h516909a_1.tar.bz2#6f8720dff19e17ce5d48cfe7f3d2f0a3
https://conda.anaconda.org/conda-forge/linux-64/libffi-3.4.2-h7f98852_5.tar.bz2#d645c6d2ac96843a2bfaccd2d62b3ac3
https://conda.anaconda.org/conda-forge/linux-64/libiconv-1.17-h166bdaf_0.tar.bz2#b62b52da46c39ee2bc3c162ac7f1804d
@@ -48,17 +49,17 @@ https://conda.anaconda.org/conda-forge/linux-64/libnsl-2.0.0-h7f98852_0.tar.bz2#
https://conda.anaconda.org/conda-forge/linux-64/libogg-1.3.4-h7f98852_1.tar.bz2#6e8cc2173440d77708196c5b93771680
https://conda.anaconda.org/conda-forge/linux-64/libopenblas-0.3.21-pthreads_h78a6416_3.tar.bz2#8c5963a49b6035c40646a763293fbb35
https://conda.anaconda.org/conda-forge/linux-64/libopus-1.3.1-h7f98852_1.tar.bz2#15345e56d527b330e1cacbdf58676e8f
-https://conda.anaconda.org/conda-forge/linux-64/libtool-2.4.6-h9c3ff4c_1008.tar.bz2#16e143a1ed4b4fd169536373957f6fee
-https://conda.anaconda.org/conda-forge/linux-64/libudev1-252-h166bdaf_0.tar.bz2#174243089ec111479298a5b7099b64b5
+https://conda.anaconda.org/conda-forge/linux-64/libtool-2.4.7-h27087fc_0.conda#f204c8ba400ec475452737094fb81d52
+https://conda.anaconda.org/conda-forge/linux-64/libudev1-253-h0b41bf4_0.conda#6c2addbd9aa4ee47c76d50c9f0df8cd6
https://conda.anaconda.org/conda-forge/linux-64/libuuid-2.32.1-h7f98852_1000.tar.bz2#772d69f030955d9646d3d0eaf21d859d
https://conda.anaconda.org/conda-forge/linux-64/libwebp-base-1.2.4-h166bdaf_0.tar.bz2#ac2ccf7323d21f2994e4d1f5da664f37
https://conda.anaconda.org/conda-forge/linux-64/libzlib-1.2.13-h166bdaf_4.tar.bz2#f3f9de449d32ca9b9c66a22863c96f41
-https://conda.anaconda.org/conda-forge/linux-64/lz4-c-1.9.3-h9c3ff4c_1.tar.bz2#fbe97e8fa6f275d7c76a09e795adc3e6
-https://conda.anaconda.org/conda-forge/linux-64/mpg123-1.31.1-h27087fc_0.tar.bz2#0af513b75f78a701a152568a31303bdf
+https://conda.anaconda.org/conda-forge/linux-64/lz4-c-1.9.4-hcb278e6_0.conda#318b08df404f9c9be5712aaa5a6f0bb0
+https://conda.anaconda.org/conda-forge/linux-64/mpg123-1.31.2-hcb278e6_0.conda#08efb1e1813f1a151b7a945b972a049b
https://conda.anaconda.org/conda-forge/linux-64/mpich-4.0.3-h846660c_100.tar.bz2#50d66bb751cfa71ee2a48b2d3eb90ac1
https://conda.anaconda.org/conda-forge/linux-64/ncurses-6.3-h27087fc_1.tar.bz2#4acfc691e64342b9dae57cf2adc63238
https://conda.anaconda.org/conda-forge/linux-64/nspr-4.35-h27087fc_0.conda#da0ec11a6454ae19bff5b02ed881a2b1
-https://conda.anaconda.org/conda-forge/linux-64/openssl-3.0.7-h0b41bf4_1.conda#7adaac6ff98219bcb99b45e408b80f4e
+https://conda.anaconda.org/conda-forge/linux-64/openssl-3.0.8-h0b41bf4_0.conda#e043403cd18faf815bf7705ab6c1e092
https://conda.anaconda.org/conda-forge/linux-64/pixman-0.40.0-h36c2ea0_0.tar.bz2#660e72c82f2e75a6b3fe6a6e75c79f19
https://conda.anaconda.org/conda-forge/linux-64/pthread-stubs-0.4-h36c2ea0_1001.tar.bz2#22dad4df6e8630e8dff2428f6f6a7036
https://conda.anaconda.org/conda-forge/linux-64/xorg-kbproto-1.0.7-h7f98852_1002.tar.bz2#4b230e8381279d76131116660f5a241a
@@ -66,12 +67,12 @@ https://conda.anaconda.org/conda-forge/linux-64/xorg-libice-1.0.10-h7f98852_0.ta
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxau-1.0.9-h7f98852_0.tar.bz2#bf6f803a544f26ebbdc3bfff272eb179
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxdmcp-1.1.3-h7f98852_0.tar.bz2#be93aabceefa2fac576e971aef407908
https://conda.anaconda.org/conda-forge/linux-64/xorg-renderproto-0.11.1-h7f98852_1002.tar.bz2#06feff3d2634e3097ce2fe681474b534
-https://conda.anaconda.org/conda-forge/linux-64/xorg-xextproto-7.3.0-h7f98852_1002.tar.bz2#1e15f6ad85a7d743a2ac68dae6c82b98
+https://conda.anaconda.org/conda-forge/linux-64/xorg-xextproto-7.3.0-h0b41bf4_1003.conda#bce9f945da8ad2ae9b1d7165a64d0f87
https://conda.anaconda.org/conda-forge/linux-64/xorg-xproto-7.0.31-h7f98852_1007.tar.bz2#b4a4381d54784606820704f7b5f05a15
-https://conda.anaconda.org/conda-forge/linux-64/xxhash-0.8.0-h7f98852_3.tar.bz2#52402c791f35e414e704b7a113f99605
+https://conda.anaconda.org/conda-forge/linux-64/xxhash-0.8.1-h0b41bf4_0.conda#e9c3bcf0e0c719431abec8ca447eee27
https://conda.anaconda.org/conda-forge/linux-64/xz-5.2.6-h166bdaf_0.tar.bz2#2161070d867d1b1204ea749c8eec4ef0
https://conda.anaconda.org/conda-forge/linux-64/yaml-0.2.5-h7f98852_2.tar.bz2#4cb3ad778ec2d5a7acbdf254eb1c42ae
-https://conda.anaconda.org/conda-forge/linux-64/jack-1.9.21-h583fa2b_2.conda#7b36a10b58964d4444fcba44244710c5
+https://conda.anaconda.org/conda-forge/linux-64/jack-1.9.22-h11f4161_0.conda#504fa9e712b99494a9cf4630e3ca7d78
https://conda.anaconda.org/conda-forge/linux-64/libblas-3.9.0-16_linux64_openblas.tar.bz2#d9b7a8639171f6c6fa0a983edabcfe2b
https://conda.anaconda.org/conda-forge/linux-64/libbrotlidec-1.0.9-h166bdaf_8.tar.bz2#4ae4d7795d33e02bd20f6b23d91caf82
https://conda.anaconda.org/conda-forge/linux-64/libbrotlienc-1.0.9-h166bdaf_8.tar.bz2#04bac51ba35ea023dc48af73c1c88c25
@@ -79,8 +80,8 @@ https://conda.anaconda.org/conda-forge/linux-64/libcap-2.66-ha37c62d_0.tar.bz2#2
https://conda.anaconda.org/conda-forge/linux-64/libedit-3.1.20191231-he28a2e2_2.tar.bz2#4d331e44109e3f0e19b4cb8f9b82f3e1
https://conda.anaconda.org/conda-forge/linux-64/libevent-2.1.10-h28343ad_4.tar.bz2#4a049fc560e00e43151dc51368915fdd
https://conda.anaconda.org/conda-forge/linux-64/libflac-1.4.2-h27087fc_0.tar.bz2#7daf72d8e2a8e848e11d63ed6d1026e0
-https://conda.anaconda.org/conda-forge/linux-64/libgpg-error-1.45-hc0c96e0_0.tar.bz2#839aeb24ab885a7b902247a6d943d02f
-https://conda.anaconda.org/conda-forge/linux-64/libnghttp2-1.47.0-hff17c54_1.tar.bz2#2b7dbfa6988a41f9d23ba6d4f0e1d74e
+https://conda.anaconda.org/conda-forge/linux-64/libgpg-error-1.46-h620e276_0.conda#27e745f6f2e4b757e95dd7225fbe6bdb
+https://conda.anaconda.org/conda-forge/linux-64/libnghttp2-1.52.0-h61bc06f_0.conda#613955a50485812985c059e7b269f42e
https://conda.anaconda.org/conda-forge/linux-64/libpng-1.6.39-h753d276_0.conda#e1c890aebdebbfbf87e2c917187b4416
https://conda.anaconda.org/conda-forge/linux-64/libsqlite-3.40.0-h753d276_0.tar.bz2#2e5f9a37d487e1019fd4d8113adb2f9f
https://conda.anaconda.org/conda-forge/linux-64/libssh2-1.10.0-hf14f497_3.tar.bz2#d85acad4b47dff4e3def14a769a97906
@@ -88,78 +89,77 @@ https://conda.anaconda.org/conda-forge/linux-64/libvorbis-1.3.7-h9c3ff4c_0.tar.b
https://conda.anaconda.org/conda-forge/linux-64/libxcb-1.13-h7f98852_1004.tar.bz2#b3653fdc58d03face9724f602218a904
https://conda.anaconda.org/conda-forge/linux-64/libxml2-2.10.3-h7463322_0.tar.bz2#3b933ea47ef8f330c4c068af25fcd6a8
https://conda.anaconda.org/conda-forge/linux-64/libzip-1.9.2-hc929e4a_1.tar.bz2#5b122b50e738c4be5c3f2899f010d7cf
-https://conda.anaconda.org/conda-forge/linux-64/mysql-common-8.0.31-h26416b9_0.tar.bz2#6c531bc30d49ae75b9c7c7f65bd62e3c
+https://conda.anaconda.org/conda-forge/linux-64/mysql-common-8.0.32-ha901b37_0.conda#6a39818710235826181e104aada40c75
https://conda.anaconda.org/conda-forge/linux-64/pcre2-10.40-hc3806b6_0.tar.bz2#69e2c796349cd9b273890bee0febfe1b
https://conda.anaconda.org/conda-forge/linux-64/readline-8.1.2-h0f457ee_0.tar.bz2#db2ebbe2943aae81ed051a6a9af8e0fa
https://conda.anaconda.org/conda-forge/linux-64/tk-8.6.12-h27826a3_0.tar.bz2#5b8c42eb62e9fc961af70bdd6a26e168
https://conda.anaconda.org/conda-forge/linux-64/udunits2-2.2.28-hc3e0081_0.tar.bz2#d4c341e0379c31e9e781d4f204726867
https://conda.anaconda.org/conda-forge/linux-64/xorg-libsm-1.2.3-hd9c2040_1000.tar.bz2#9e856f78d5c80d5a78f61e72d1d473a3
https://conda.anaconda.org/conda-forge/linux-64/zlib-1.2.13-h166bdaf_4.tar.bz2#4b11e365c0275b808be78b30f904e295
-https://conda.anaconda.org/conda-forge/linux-64/zstd-1.5.2-h6239696_4.tar.bz2#adcf0be7897e73e312bd24353b613f74
+https://conda.anaconda.org/conda-forge/linux-64/zstd-1.5.2-h3eb15da_6.conda#6b63daed8feeca47be78f323e793d555
https://conda.anaconda.org/conda-forge/linux-64/brotli-bin-1.0.9-h166bdaf_8.tar.bz2#e5613f2bc717e9945840ff474419b8e4
https://conda.anaconda.org/conda-forge/linux-64/freetype-2.12.1-hca18f0e_1.conda#e1232042de76d24539a436d37597eb06
https://conda.anaconda.org/conda-forge/linux-64/hdf4-4.2.15-h9772cbc_5.tar.bz2#ee08782aff2ff9b3291c967fa6bc7336
-https://conda.anaconda.org/conda-forge/linux-64/krb5-1.19.3-h08a2579_0.tar.bz2#d25e05e7ee0e302b52d24491db4891eb
+https://conda.anaconda.org/conda-forge/linux-64/krb5-1.20.1-h81ceb04_0.conda#89a41adce7106749573d883b2f657d78
https://conda.anaconda.org/conda-forge/linux-64/libcblas-3.9.0-16_linux64_openblas.tar.bz2#20bae26d0a1db73f758fc3754cab4719
https://conda.anaconda.org/conda-forge/linux-64/libgcrypt-1.10.1-h166bdaf_0.tar.bz2#f967fc95089cd247ceed56eda31de3a9
https://conda.anaconda.org/conda-forge/linux-64/libglib-2.74.1-h606061b_1.tar.bz2#ed5349aa96776e00b34eccecf4a948fe
https://conda.anaconda.org/conda-forge/linux-64/liblapack-3.9.0-16_linux64_openblas.tar.bz2#955d993f41f9354bf753d29864ea20ad
-https://conda.anaconda.org/conda-forge/linux-64/libllvm15-15.0.6-h63197d8_0.conda#201168ef66095bbd565e124ee2c56a20
-https://conda.anaconda.org/conda-forge/linux-64/libsndfile-1.1.0-hcb278e6_1.conda#d7a07b1f5974bce4735112aaef0c1467
-https://conda.anaconda.org/conda-forge/linux-64/libtiff-4.4.0-h55922b4_4.tar.bz2#901791f0ec7cddc8714e76e273013a91
-https://conda.anaconda.org/conda-forge/linux-64/libxkbcommon-1.0.3-he3ba5ed_0.tar.bz2#f9dbabc7e01c459ed7a1d1d64b206e9b
-https://conda.anaconda.org/conda-forge/linux-64/mysql-libs-8.0.31-hbc51c84_0.tar.bz2#da9633eee814d4e910fe42643a356315
-https://conda.anaconda.org/conda-forge/linux-64/nss-3.82-he02c5a1_0.conda#f8d7f11d19e4cb2207eab159fd4c0152
-https://conda.anaconda.org/conda-forge/linux-64/python-3.10.8-h4a9ceb5_0_cpython.conda#be2a6d78752c2ab85f360ce37d2c64e2
+https://conda.anaconda.org/conda-forge/linux-64/libllvm15-15.0.7-hadd5161_0.conda#70cbb0c2033665f2a7339bf0ec51a67f
+https://conda.anaconda.org/conda-forge/linux-64/libsndfile-1.2.0-hb75c966_0.conda#c648d19cd9c8625898d5d370414de7c7
+https://conda.anaconda.org/conda-forge/linux-64/libtiff-4.5.0-h6adf6a1_2.conda#2e648a34072eb39d7c4fc2a9981c5f0c
+https://conda.anaconda.org/conda-forge/linux-64/libxkbcommon-1.5.0-h79f4944_0.conda#3f67368c9b0e77a693acad193310baf1
+https://conda.anaconda.org/conda-forge/linux-64/mysql-libs-8.0.32-hd7da12d_0.conda#b05d7ea8b76f1172d5fe4f30e03277ea
+https://conda.anaconda.org/conda-forge/linux-64/nss-3.88-he45b914_0.conda#d7a81dfb99ad8fbb88872fb7ec646e6c
+https://conda.anaconda.org/conda-forge/linux-64/python-3.10.9-he550d4f_0_cpython.conda#3cb3e91b3fe66baa68a12c85f39b9b40
https://conda.anaconda.org/conda-forge/linux-64/sqlite-3.40.0-h4ff8645_0.tar.bz2#bb11803129cbbb53ed56f9506ff74145
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-0.4.0-h166bdaf_0.tar.bz2#384e7fcb3cd162ba3e4aed4b687df566
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-keysyms-0.4.0-h166bdaf_0.tar.bz2#637054603bb7594302e3bf83f0a99879
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-renderutil-0.3.9-h166bdaf_0.tar.bz2#732e22f1741bccea861f5668cf7342a7
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-wm-0.4.1-h166bdaf_0.tar.bz2#0a8e20a8aef954390b9481a527421a8c
-https://conda.anaconda.org/conda-forge/linux-64/xorg-libx11-1.7.2-h7f98852_0.tar.bz2#12a61e640b8894504326aadafccbb790
-https://conda.anaconda.org/conda-forge/noarch/alabaster-0.7.12-py_0.tar.bz2#2489a97287f90176ecdc3ca982b4b0a0
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libx11-1.8.4-h0b41bf4_0.conda#ea8fbfeb976ac49cbeb594e985393514
+https://conda.anaconda.org/conda-forge/noarch/alabaster-0.7.13-pyhd8ed1ab_0.conda#06006184e203b61d3525f90de394471e
https://conda.anaconda.org/conda-forge/linux-64/antlr-python-runtime-4.7.2-py310hff52083_1003.tar.bz2#8324f8fff866055d4b32eb25e091fe31
https://conda.anaconda.org/conda-forge/linux-64/atk-1.0-2.38.0-hd4edc92_1.tar.bz2#6c72ec3e660a51736913ef6ea68c454b
-https://conda.anaconda.org/conda-forge/noarch/attrs-22.1.0-pyh71513ae_1.tar.bz2#6d3ccbc56256204925bfa8378722792f
+https://conda.anaconda.org/conda-forge/noarch/attrs-22.2.0-pyh71513ae_0.conda#8b76db7818a4e401ed4486c4c1635cd9
https://conda.anaconda.org/conda-forge/linux-64/brotli-1.0.9-h166bdaf_8.tar.bz2#2ff08978892a3e8b954397c461f18418
https://conda.anaconda.org/conda-forge/noarch/certifi-2022.12.7-pyhd8ed1ab_0.conda#fb9addc3db06e56abe03e0e9f21a63e6
https://conda.anaconda.org/conda-forge/noarch/cfgv-3.3.1-pyhd8ed1ab_0.tar.bz2#ebb5f5f7dc4f1a3780ef7ea7738db08c
https://conda.anaconda.org/conda-forge/noarch/charset-normalizer-2.1.1-pyhd8ed1ab_0.tar.bz2#c1d5b294fbf9a795dec349a6f4d8be8e
https://conda.anaconda.org/conda-forge/noarch/click-8.1.3-unix_pyhd8ed1ab_2.tar.bz2#20e4087407c7cb04a40817114b333dbf
-https://conda.anaconda.org/conda-forge/noarch/cloudpickle-2.2.0-pyhd8ed1ab_0.tar.bz2#a6cf47b09786423200d7982d1faa19eb
+https://conda.anaconda.org/conda-forge/noarch/cloudpickle-2.2.1-pyhd8ed1ab_0.conda#b325bfc4cff7d7f8a868f1f7ecc4ed16
https://conda.anaconda.org/conda-forge/noarch/colorama-0.4.6-pyhd8ed1ab_0.tar.bz2#3faab06a954c2a04039983f2c4a50d99
https://conda.anaconda.org/conda-forge/noarch/cycler-0.11.0-pyhd8ed1ab_0.tar.bz2#a50559fad0affdbb33729a68669ca1cb
https://conda.anaconda.org/conda-forge/linux-64/dbus-1.13.6-h5008d03_3.tar.bz2#ecfff944ba3960ecb334b9a2663d708d
https://conda.anaconda.org/conda-forge/noarch/distlib-0.3.6-pyhd8ed1ab_0.tar.bz2#b65b4d50dbd2d50fa0aeac367ec9eed7
https://conda.anaconda.org/conda-forge/linux-64/docutils-0.17.1-py310hff52083_3.tar.bz2#785160da087cf1d70e989afbb761f01c
-https://conda.anaconda.org/conda-forge/noarch/exceptiongroup-1.0.4-pyhd8ed1ab_0.tar.bz2#e0734d1f12de77f9daca98bda3428733
+https://conda.anaconda.org/conda-forge/noarch/exceptiongroup-1.1.0-pyhd8ed1ab_0.conda#a385c3e8968b4cf8fbc426ace915fd1a
https://conda.anaconda.org/conda-forge/noarch/execnet-1.9.0-pyhd8ed1ab_0.tar.bz2#0e521f7a5e60d508b121d38b04874fb2
-https://conda.anaconda.org/conda-forge/noarch/filelock-3.8.2-pyhd8ed1ab_0.conda#0f09c2bc17ddd8732be8e5b99297c7ce
-https://conda.anaconda.org/conda-forge/linux-64/fontconfig-2.14.1-hc2a2eb6_0.tar.bz2#78415f0180a8d9c5bcc47889e00d5fb1
-https://conda.anaconda.org/conda-forge/noarch/fsspec-2022.11.0-pyhd8ed1ab_0.tar.bz2#eb919f2119a6db5d0192f9e9c3711572
-https://conda.anaconda.org/conda-forge/linux-64/gdk-pixbuf-2.42.8-hff1cb4f_1.tar.bz2#a61c6312192e7c9de71548a6706a21e6
+https://conda.anaconda.org/conda-forge/noarch/filelock-3.9.0-pyhd8ed1ab_0.conda#1addc115923d646ca19ed90edc413506
+https://conda.anaconda.org/conda-forge/linux-64/fontconfig-2.14.2-h14ed4e7_0.conda#0f69b688f52ff6da70bccb7ff7001d1d
+https://conda.anaconda.org/conda-forge/noarch/fsspec-2023.1.0-pyhd8ed1ab_0.conda#44f6828b8f7cc3433d68d1d1c0e9add2
+https://conda.anaconda.org/conda-forge/linux-64/gdk-pixbuf-2.42.10-h05c8ddd_0.conda#1a109126a43003d65b39c1cad656bc9b
https://conda.anaconda.org/conda-forge/linux-64/glib-tools-2.74.1-h6239696_1.tar.bz2#5f442e6bc9d89ba236eb25a25c5c2815
https://conda.anaconda.org/conda-forge/linux-64/gts-0.7.6-h64030ff_2.tar.bz2#112eb9b5b93f0c02e59aea4fd1967363
https://conda.anaconda.org/conda-forge/noarch/idna-3.4-pyhd8ed1ab_0.tar.bz2#34272b248891bddccc64479f9a7fffed
https://conda.anaconda.org/conda-forge/noarch/imagesize-1.4.1-pyhd8ed1ab_0.tar.bz2#7de5386c8fea29e76b303f37dde4c352
-https://conda.anaconda.org/conda-forge/noarch/iniconfig-1.1.1-pyh9f0ad1d_0.tar.bz2#39161f81cc5e5ca45b8226fbb06c6905
+https://conda.anaconda.org/conda-forge/noarch/iniconfig-2.0.0-pyhd8ed1ab_0.conda#f800d2da156d08e289b14e87e43c1ae5
https://conda.anaconda.org/conda-forge/noarch/iris-sample-data-2.4.0-pyhd8ed1ab_0.tar.bz2#18ee9c07cf945a33f92caf1ee3d23ad9
https://conda.anaconda.org/conda-forge/linux-64/kiwisolver-1.4.4-py310hbf28c38_1.tar.bz2#ad5647e517ba68e2868ef2e6e6ff7723
-https://conda.anaconda.org/conda-forge/linux-64/lcms2-2.14-h6ed2654_0.tar.bz2#dcc588839de1445d90995a0a2c4f3a39
-https://conda.anaconda.org/conda-forge/linux-64/libclang13-15.0.6-default_h3a83d3e_0.conda#535dd0ca1dcb165b6a8ffa10d01945fe
-https://conda.anaconda.org/conda-forge/linux-64/libcups-2.3.3-h3e49a29_2.tar.bz2#3b88f1d0fe2580594d58d7e44d664617
-https://conda.anaconda.org/conda-forge/linux-64/libcurl-7.86.0-h2283fc2_1.tar.bz2#fdca8cd67ec2676f90a70ac73a32538b
-https://conda.anaconda.org/conda-forge/linux-64/libpq-15.1-h67c24c5_1.conda#e1389a8d9a907133b3e6483c2807d243
+https://conda.anaconda.org/conda-forge/linux-64/lcms2-2.14-hfd0df8a_1.conda#c2566c2ea5f153ddd6bf4acaf7547d97
+https://conda.anaconda.org/conda-forge/linux-64/libclang13-15.0.7-default_h3e3d535_1.conda#a3a0f7a6f0885f5e1e0ec691566afb77
+https://conda.anaconda.org/conda-forge/linux-64/libcups-2.3.3-h36d4200_3.conda#c9f4416a34bc91e0eb029f912c68f81f
+https://conda.anaconda.org/conda-forge/linux-64/libcurl-7.88.1-hdc1c0ab_0.conda#81eaeb3b35163c8e90e57532bc93754d
+https://conda.anaconda.org/conda-forge/linux-64/libpq-15.2-hb675445_0.conda#4654b17eccaba55b8581d6b9c77f53cc
https://conda.anaconda.org/conda-forge/linux-64/libsystemd0-252-h2a991cd_0.tar.bz2#3c5ae9f61f663b3d5e1bf7f7da0c85f5
-https://conda.anaconda.org/conda-forge/linux-64/libwebp-1.2.4-h522a892_0.tar.bz2#802e43f480122a85ae6a34c1909f8f98
+https://conda.anaconda.org/conda-forge/linux-64/libwebp-1.2.4-h1daa5a0_1.conda#77003f63d1763c1e6569a02c1742c9f4
https://conda.anaconda.org/conda-forge/noarch/locket-1.0.0-pyhd8ed1ab_0.tar.bz2#91e27ef3d05cc772ce627e51cff111c4
-https://conda.anaconda.org/conda-forge/linux-64/markupsafe-2.1.1-py310h5764c6d_2.tar.bz2#2d7028ea2a77f909931e1a173d952261
+https://conda.anaconda.org/conda-forge/linux-64/markupsafe-2.1.2-py310h1fa729e_0.conda#a1f0db6709778b77b5903541eeac4032
https://conda.anaconda.org/conda-forge/linux-64/mpi4py-3.1.4-py310h37cc914_0.tar.bz2#98d598d9178d7f3091212c61c0be693c
https://conda.anaconda.org/conda-forge/noarch/munkres-1.1.4-pyh9f0ad1d_0.tar.bz2#2ba8498c1018c1e9c61eb99b973dfe19
-https://conda.anaconda.org/conda-forge/linux-64/numpy-1.23.5-py310h53a5b5f_0.conda#3b114b1559def8bad228fec544ac1812
-https://conda.anaconda.org/conda-forge/linux-64/openjpeg-2.5.0-h7d73246_1.tar.bz2#a11b4df9271a8d7917686725aa04c8f2
-https://conda.anaconda.org/conda-forge/noarch/packaging-22.0-pyhd8ed1ab_0.conda#0e8e1bd93998978fc3125522266d12db
-https://conda.anaconda.org/conda-forge/noarch/platformdirs-2.6.0-pyhd8ed1ab_0.conda#b1b2ab02d1ece1719f7fa002ad4bc70d
+https://conda.anaconda.org/conda-forge/linux-64/numpy-1.24.2-py310h8deb116_0.conda#b7085457309e206174b8e234d90a7605
+https://conda.anaconda.org/conda-forge/linux-64/openjpeg-2.5.0-hfec8fc6_2.conda#5ce6a42505c6e9e6151c54c3ec8d68ea
+https://conda.anaconda.org/conda-forge/noarch/packaging-23.0-pyhd8ed1ab_0.conda#1ff2e3ca41f0ce16afec7190db28288b
https://conda.anaconda.org/conda-forge/noarch/pluggy-1.0.0-pyhd8ed1ab_5.tar.bz2#7d301a0d25f424d96175f810935f0da9
https://conda.anaconda.org/conda-forge/noarch/ply-3.11-py_1.tar.bz2#7205635cd71531943440fbfe3b6b5727
https://conda.anaconda.org/conda-forge/linux-64/psutil-5.9.4-py310h5764c6d_0.tar.bz2#c3c55664e9becc48e6a652e2b641961f
@@ -167,16 +167,16 @@ https://conda.anaconda.org/conda-forge/noarch/pycparser-2.21-pyhd8ed1ab_0.tar.bz
https://conda.anaconda.org/conda-forge/noarch/pyparsing-3.0.9-pyhd8ed1ab_0.tar.bz2#e8fbc1b54b25f4b08281467bc13b70cc
https://conda.anaconda.org/conda-forge/noarch/pyshp-2.3.1-pyhd8ed1ab_0.tar.bz2#92a889dc236a5197612bc85bee6d7174
https://conda.anaconda.org/conda-forge/noarch/pysocks-1.7.1-pyha2e5f31_6.tar.bz2#2a7de29fb590ca14b5243c4c812c8025
-https://conda.anaconda.org/conda-forge/linux-64/python-xxhash-3.0.0-py310h5764c6d_2.tar.bz2#cce72b32ccc346ed166fc85071854a86
-https://conda.anaconda.org/conda-forge/noarch/pytz-2022.6-pyhd8ed1ab_0.tar.bz2#b1f26ad83328e486910ef7f6e81dc061
+https://conda.anaconda.org/conda-forge/linux-64/python-xxhash-3.2.0-py310h1fa729e_0.conda#8d155ac95b1dfe585bcb6bec6a91c73b
+https://conda.anaconda.org/conda-forge/noarch/pytz-2022.7.1-pyhd8ed1ab_0.conda#f59d49a7b464901cf714b9e7984d01a2
https://conda.anaconda.org/conda-forge/linux-64/pyyaml-6.0-py310h5764c6d_5.tar.bz2#9e68d2ff6d98737c855b65f48dd3c597
-https://conda.anaconda.org/conda-forge/noarch/setuptools-65.5.1-pyhd8ed1ab_0.tar.bz2#cfb8dc4d9d285ca5fb1177b9dd450e33
+https://conda.anaconda.org/conda-forge/noarch/setuptools-67.4.0-pyhd8ed1ab_0.conda#c6f4b87020c72e2700e3e94c1fc93b70
https://conda.anaconda.org/conda-forge/noarch/six-1.16.0-pyh6c4a22f_0.tar.bz2#e5f25f8dbc060e9a8d912e432202afc2
https://conda.anaconda.org/conda-forge/noarch/snowballstemmer-2.2.0-pyhd8ed1ab_0.tar.bz2#4d22a9315e78c6827f806065957d566e
https://conda.anaconda.org/conda-forge/noarch/soupsieve-2.3.2.post1-pyhd8ed1ab_0.tar.bz2#146f4541d643d48fc8a75cacf69f03ae
-https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-applehelp-1.0.2-py_0.tar.bz2#20b2eaeaeea4ef9a9a0d99770620fd09
+https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-applehelp-1.0.4-pyhd8ed1ab_0.conda#5a31a7d564f551d0e6dff52fd8cb5b16
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-devhelp-1.0.2-py_0.tar.bz2#68e01cac9d38d0e717cd5c87bc3d2cc9
-https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-htmlhelp-2.0.0-pyhd8ed1ab_0.tar.bz2#77dad82eb9c8c1525ff7953e0756d708
+https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-htmlhelp-2.0.1-pyhd8ed1ab_0.conda#6c8c4d6eb2325e59290ac6dbbeacd5f0
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-jsmath-1.0.1-py_0.tar.bz2#67cd9d9c0382d37479b4d306c369a2d4
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-qthelp-1.0.3-py_0.tar.bz2#d01180388e6d1838c3e1ad029590aa7a
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-serializinghtml-1.1.5-pyhd8ed1ab_2.tar.bz2#9ff55a0901cf952f05c654394de76bf7
@@ -188,78 +188,84 @@ https://conda.anaconda.org/conda-forge/noarch/typing_extensions-4.4.0-pyha770c72
https://conda.anaconda.org/conda-forge/linux-64/unicodedata2-15.0.0-py310h5764c6d_0.tar.bz2#e972c5a1f472561cf4a91962cb01f4b4
https://conda.anaconda.org/conda-forge/noarch/wheel-0.38.4-pyhd8ed1ab_0.tar.bz2#c829cfb8cb826acb9de0ac1a2df0a940
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-image-0.4.0-h166bdaf_0.tar.bz2#c9b568bd804cb2903c6be6f5f68182e4
-https://conda.anaconda.org/conda-forge/linux-64/xorg-libxext-1.3.4-h7f98852_1.tar.bz2#536cc5db4d0a3ba0630541aec064b5e4
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxext-1.3.4-h0b41bf4_2.conda#82b6df12252e6f32402b96dacc656fec
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxrender-0.9.10-h7f98852_1003.tar.bz2#f59c1242cc1dd93e72c2ee2b360979eb
-https://conda.anaconda.org/conda-forge/noarch/zipp-3.11.0-pyhd8ed1ab_0.conda#09b5b885341697137879a4f039a9e5a1
-https://conda.anaconda.org/conda-forge/noarch/babel-2.11.0-pyhd8ed1ab_0.tar.bz2#2ea70fde8d581ba9425a761609eed6ba
-https://conda.anaconda.org/conda-forge/noarch/beautifulsoup4-4.11.1-pyha770c72_0.tar.bz2#eeec8814bd97b2681f708bb127478d7d
+https://conda.anaconda.org/conda-forge/noarch/zipp-3.15.0-pyhd8ed1ab_0.conda#13018819ca8f5b7cc675a8faf1f5fedf
+https://conda.anaconda.org/conda-forge/noarch/babel-2.12.1-pyhd8ed1ab_1.conda#ac432e732804a81ddcf29c92ead57cde
+https://conda.anaconda.org/conda-forge/noarch/beautifulsoup4-4.11.2-pyha770c72_0.conda#88b59f6989f0ed5ab3433af0b82555e1
https://conda.anaconda.org/conda-forge/linux-64/cairo-1.16.0-ha61ee94_1014.tar.bz2#d1a88f3ed5b52e1024b80d4bcd26a7a0
-https://conda.anaconda.org/conda-forge/linux-64/cffi-1.15.1-py310h255011f_2.tar.bz2#6bb8063dd08f9724c18744b0e040cfe2
+https://conda.anaconda.org/conda-forge/linux-64/cffi-1.15.1-py310h255011f_3.conda#800596144bb613cd7ac58b80900ce835
https://conda.anaconda.org/conda-forge/linux-64/cftime-1.6.2-py310hde88566_1.tar.bz2#94ce7a76b0c912279f6958e0b6b21d2b
-https://conda.anaconda.org/conda-forge/linux-64/contourpy-1.0.6-py310hbf28c38_0.tar.bz2#c5b1699e390d30b680dd93a2b251062b
-https://conda.anaconda.org/conda-forge/linux-64/curl-7.86.0-h2283fc2_1.tar.bz2#9d4149760567cb232691cce2d8ccc21f
+https://conda.anaconda.org/conda-forge/linux-64/contourpy-1.0.7-py310hdf3cbec_0.conda#7bf9d8c765b6b04882c719509652c6d6
+https://conda.anaconda.org/conda-forge/linux-64/coverage-7.2.1-py310h1fa729e_0.conda#42814f7e7ce2e3e7d048c4efea481759
+https://conda.anaconda.org/conda-forge/linux-64/curl-7.88.1-hdc1c0ab_0.conda#1968e4fef727858ac04746560e820928
https://conda.anaconda.org/conda-forge/linux-64/fonttools-4.38.0-py310h5764c6d_1.tar.bz2#12ebe92a8a578bc903bd844744f4d040
https://conda.anaconda.org/conda-forge/linux-64/glib-2.74.1-h6239696_1.tar.bz2#f3220a9e9d3abcbfca43419a219df7e4
-https://conda.anaconda.org/conda-forge/linux-64/hdf5-1.12.2-mpi_mpich_h5d83325_0.tar.bz2#6b5c2d276f306df759cfbdb0f41c4db9
-https://conda.anaconda.org/conda-forge/noarch/importlib-metadata-5.1.0-pyha770c72_0.conda#46a62e35b9ae515cf0e49afc7fe0e7ef
+https://conda.anaconda.org/conda-forge/linux-64/hdf5-1.12.2-mpi_mpich_h5d83325_1.conda#811c4d55cf17b42336ffa314239717b0
+https://conda.anaconda.org/conda-forge/noarch/importlib-metadata-6.0.0-pyha770c72_0.conda#691644becbcdca9f73243450b1c63e62
https://conda.anaconda.org/conda-forge/noarch/jinja2-3.1.2-pyhd8ed1ab_1.tar.bz2#c8490ed5c70966d232fdd389d0dbed37
-https://conda.anaconda.org/conda-forge/linux-64/libclang-15.0.6-default_h2e3cab8_0.conda#1b2cee49acc5b03c73ad0f68bfe04bb8
-https://conda.anaconda.org/conda-forge/linux-64/libgd-2.3.3-h18fbbfe_3.tar.bz2#ea9758cf553476ddf75c789fdd239dc5
+https://conda.anaconda.org/conda-forge/linux-64/libclang-15.0.7-default_had23c3d_1.conda#36c65ed73b7c92589bd9562ef8a6023d
+https://conda.anaconda.org/conda-forge/linux-64/libgd-2.3.3-h5aea950_4.conda#82ef57611ace65b59db35a9687264572
https://conda.anaconda.org/conda-forge/linux-64/mo_pack-0.2.0-py310hde88566_1008.tar.bz2#f9dd8a7a2fcc23eb2cd95cd817c949e7
https://conda.anaconda.org/conda-forge/noarch/nodeenv-1.7.0-pyhd8ed1ab_0.tar.bz2#fbe1182f650c04513046d6894046cd6c
https://conda.anaconda.org/conda-forge/noarch/partd-1.3.0-pyhd8ed1ab_0.tar.bz2#af8c82d121e63082926062d61d9abb54
-https://conda.anaconda.org/conda-forge/linux-64/pillow-9.2.0-py310h454ad03_3.tar.bz2#eb354ff791f505b1d6f13f776359d88e
-https://conda.anaconda.org/conda-forge/noarch/pip-22.3.1-pyhd8ed1ab_0.tar.bz2#da66f2851b9836d3a7c5190082a45f7d
+https://conda.anaconda.org/conda-forge/linux-64/pillow-9.4.0-py310h023d228_1.conda#bbea829b541aa15df5c65bd40b8c1981
+https://conda.anaconda.org/conda-forge/noarch/pip-23.0.1-pyhd8ed1ab_0.conda#8025ca83b8ba5430b640b83917c2a6f7
https://conda.anaconda.org/conda-forge/noarch/pockets-0.9.1-py_0.tar.bz2#1b52f0c42e8077e5a33e00fe72269364
-https://conda.anaconda.org/conda-forge/linux-64/proj-9.1.0-h93bde94_0.tar.bz2#255c7204dda39747c3ba380d28b026d7
-https://conda.anaconda.org/conda-forge/linux-64/pulseaudio-16.1-h126f2b6_0.tar.bz2#e4b74b33e13dd146e7d8b5078fc9ad30
-https://conda.anaconda.org/conda-forge/noarch/pygments-2.13.0-pyhd8ed1ab_0.tar.bz2#9f478e8eedd301008b5f395bad0caaed
-https://conda.anaconda.org/conda-forge/noarch/pytest-7.2.0-pyhd8ed1ab_2.tar.bz2#ac82c7aebc282e6ac0450fca012ca78c
+https://conda.anaconda.org/conda-forge/linux-64/proj-9.1.1-h8ffa02c_2.conda#c264aea0e16bba26afa0a0940e954492
+https://conda.anaconda.org/conda-forge/linux-64/pulseaudio-16.1-ha8d29e2_1.conda#dbfc2a8d63a43a11acf4c704e1ef9d0c
+https://conda.anaconda.org/conda-forge/noarch/pygments-2.14.0-pyhd8ed1ab_0.conda#c78cd16b11cd6a295484bd6c8f24bea1
+https://conda.anaconda.org/conda-forge/noarch/pytest-7.2.2-pyhd8ed1ab_0.conda#60958b19354e0ec295b43f6ab5cfab86
https://conda.anaconda.org/conda-forge/noarch/python-dateutil-2.8.2-pyhd8ed1ab_0.tar.bz2#dd999d1cc9f79e67dbb855c8924c7984
https://conda.anaconda.org/conda-forge/linux-64/python-stratify-0.2.post0-py310hde88566_3.tar.bz2#0b686f306a76fba9a61e7019f854321f
-https://conda.anaconda.org/conda-forge/linux-64/pywavelets-1.3.0-py310hde88566_2.tar.bz2#61e2f2f7befaf45f47d1da449a9a0aca
-https://conda.anaconda.org/conda-forge/linux-64/scipy-1.9.3-py310hdfbd76f_2.tar.bz2#0582a434d03f6b06d5defbb142c96f4f
-https://conda.anaconda.org/conda-forge/linux-64/shapely-1.8.5-py310h5b266fc_2.tar.bz2#c4a3707d6a630facb6cf7ed8e0d37326
-https://conda.anaconda.org/conda-forge/linux-64/sip-6.7.5-py310hd8f1fbe_0.conda#765b39936044b542a69ec2d863f5b891
+https://conda.anaconda.org/conda-forge/linux-64/pywavelets-1.4.1-py310h0a54255_0.conda#b9e952fe3f7528ab603d2776175ba8d2
+https://conda.anaconda.org/conda-forge/linux-64/shapely-2.0.1-py310h8b84c32_0.conda#965113c401c7dc9b7a4cd5f9af57e185
+https://conda.anaconda.org/conda-forge/linux-64/sip-6.7.7-py310heca2aa9_0.conda#142c074701cf90c88667b461678aee81
https://conda.anaconda.org/conda-forge/noarch/typing-extensions-4.4.0-hd8ed1ab_0.tar.bz2#be969210b61b897775a0de63cd9e9026
-https://conda.anaconda.org/conda-forge/linux-64/virtualenv-20.17.0-py310hff52083_0.conda#c6fc5e3f0a463ddb59cfda9a1582cfa0
+https://conda.anaconda.org/conda-forge/noarch/accessible-pygments-0.0.3-pyhd8ed1ab_0.conda#9838acb5f38ac58240741f5cea70a952
https://conda.anaconda.org/conda-forge/linux-64/brotlipy-0.7.0-py310h5764c6d_1005.tar.bz2#87669c3468dff637bbd0363bc0f895cf
https://conda.anaconda.org/conda-forge/linux-64/cf-units-3.1.1-py310hde88566_2.tar.bz2#7433944046deda7775c5b1f7e0b6fe18
-https://conda.anaconda.org/conda-forge/linux-64/cryptography-38.0.4-py310h600f1e7_0.conda#f999dcc21fe27ad97a8afcfa590daa14
-https://conda.anaconda.org/conda-forge/noarch/dask-core-2022.12.0-pyhd8ed1ab_0.conda#3a0f020d07998e1ae711df071f97fc19
-https://conda.anaconda.org/conda-forge/linux-64/gstreamer-1.21.2-hd4edc92_0.conda#3ae425efddb9da5fb35edda331e4dff7
-https://conda.anaconda.org/conda-forge/linux-64/harfbuzz-5.3.0-h418a68e_0.tar.bz2#888056bd4b12e110b10d4d1f29161c5e
-https://conda.anaconda.org/conda-forge/noarch/imagehash-4.3.1-pyhd8ed1ab_0.tar.bz2#132ad832787a2156be1f1b309835001a
-https://conda.anaconda.org/conda-forge/linux-64/libnetcdf-4.8.1-mpi_mpich_hcd871d9_6.tar.bz2#6cdc429ed22edb566ac4308f3da6916d
-https://conda.anaconda.org/conda-forge/linux-64/matplotlib-base-3.6.2-py310h8d5ebf3_0.tar.bz2#da51ddb20c0f99d672eb756c3abf27e7
-https://conda.anaconda.org/conda-forge/linux-64/pandas-1.5.2-py310h769672d_0.conda#bc363997d22f3b058fb17f1e89d4c96f
-https://conda.anaconda.org/conda-forge/linux-64/pyproj-3.4.0-py310hb1338dc_2.tar.bz2#e1648c222911ad7559d62831e4bc447c
-https://conda.anaconda.org/conda-forge/linux-64/pyqt5-sip-12.11.0-py310hd8f1fbe_2.tar.bz2#0d815f1b2258d3d4c17cc80fd01e0f36
-https://conda.anaconda.org/conda-forge/noarch/pytest-xdist-3.1.0-pyhd8ed1ab_0.conda#e82f8fb903d7c4a59c77954759c341f9
-https://conda.anaconda.org/conda-forge/noarch/setuptools-scm-7.0.5-pyhd8ed1ab_1.tar.bz2#07037fe2931871ed69b2b3d2acd5fdc6
+https://conda.anaconda.org/conda-forge/linux-64/cryptography-39.0.2-py310h34c0648_0.conda#99dc5a02a8b16cd88ca9a12354496e78
+https://conda.anaconda.org/conda-forge/noarch/dask-core-2023.3.0-pyhd8ed1ab_0.conda#34437340f37faafad7a6287d3b624f60
+https://conda.anaconda.org/conda-forge/linux-64/gstreamer-1.22.0-h25f0c4b_0.conda#d764367398de61c0d5531dd912e6cc96
+https://conda.anaconda.org/conda-forge/linux-64/harfbuzz-6.0.0-h8e241bc_0.conda#448fe40d2fed88ccf4d9ded37cbb2b38
+https://conda.anaconda.org/conda-forge/linux-64/libnetcdf-4.9.1-mpi_mpich_hf10a581_1.conda#1d235cbeed74dc63e22e41779838bec1
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-base-3.7.0-py310he60537e_0.conda#83a21bbd1c6fbeb339ba914fb5e5c02d
+https://conda.anaconda.org/conda-forge/linux-64/pandas-1.5.3-py310h9b08913_0.conda#467244b0dbb7da40927ac6ee0e9491de
+https://conda.anaconda.org/conda-forge/noarch/platformdirs-3.1.0-pyhd8ed1ab_0.conda#0b8fbdfd52918bc2f1b76feccd95c919
+https://conda.anaconda.org/conda-forge/linux-64/pyproj-3.4.1-py310h15e2413_1.conda#5be35366687def87437d210fd673100c
+https://conda.anaconda.org/conda-forge/linux-64/pyqt5-sip-12.11.0-py310heca2aa9_3.conda#3b1946b676534472ce65181dda0b9554
+https://conda.anaconda.org/conda-forge/noarch/pytest-cov-4.0.0-pyhd8ed1ab_0.tar.bz2#c9e3f8bfdb9bfc34aa1836a6ed4b25d7
+https://conda.anaconda.org/conda-forge/noarch/pytest-xdist-3.2.0-pyhd8ed1ab_0.conda#70ab87b96126f35d1e68de2ad9fb6423
+https://conda.anaconda.org/conda-forge/noarch/setuptools-scm-7.1.0-pyhd8ed1ab_0.conda#6613dbb3b25cc648a107f33ca9f80fc1
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-napoleon-0.7-py_0.tar.bz2#0bc25ff6f2e34af63ded59692df5f749
https://conda.anaconda.org/conda-forge/linux-64/ukkonen-1.0.1-py310hbf28c38_3.tar.bz2#703ff1ac7d1b27fb5944b8052b5d1edb
-https://conda.anaconda.org/conda-forge/linux-64/cartopy-0.21.0-py310h83f2385_3.tar.bz2#4ec35f7eebe4221c1c00fdd6540db4dc
-https://conda.anaconda.org/conda-forge/linux-64/gst-plugins-base-1.21.2-h3e40eee_0.conda#52cbed7e92713cf01b76445530396695
-https://conda.anaconda.org/conda-forge/noarch/identify-2.5.9-pyhd8ed1ab_0.conda#e7ecbbb61a37daed2a13de43d35d5282
+https://conda.anaconda.org/conda-forge/linux-64/gst-plugins-base-1.22.0-h4243ec0_0.conda#81c20b15d2281a1ea48eac5b4eee8cfa
+https://conda.anaconda.org/conda-forge/noarch/identify-2.5.18-pyhd8ed1ab_0.conda#e07a5691c27e65d8d3d9278c578c7771
https://conda.anaconda.org/conda-forge/noarch/nc-time-axis-1.4.1-pyhd8ed1ab_0.tar.bz2#281b58948bf60a2582de9e548bcc5369
-https://conda.anaconda.org/conda-forge/linux-64/netcdf-fortran-4.6.0-mpi_mpich_hd09bd1e_1.tar.bz2#0b69750bb937cab0db14f6bcef6fd787
-https://conda.anaconda.org/conda-forge/linux-64/netcdf4-1.6.0-nompi_py310h0a86a1f_103.conda#7f69695b684f2595d9ba1ce26d693b7d
-https://conda.anaconda.org/conda-forge/linux-64/pango-1.50.12-h382ae3d_0.conda#627bea5af786dbd8013ef26127d8115a
-https://conda.anaconda.org/conda-forge/noarch/pyopenssl-22.1.0-pyhd8ed1ab_0.tar.bz2#fbfa0a180d48c800f922a10a114a8632
-https://conda.anaconda.org/conda-forge/linux-64/esmf-8.2.0-mpi_mpich_h5a1934d_102.tar.bz2#bb8bdfa5e3e9e3f6ec861f05cd2ad441
+https://conda.anaconda.org/conda-forge/linux-64/netcdf-fortran-4.6.0-mpi_mpich_ha3603da_3.conda#ea38e2d3c472876ff4bf6551c17a9a1a
+https://conda.anaconda.org/conda-forge/linux-64/netcdf4-1.6.2-nompi_py310h0feb132_101.conda#acac875fc7bd759386277c6bca56b064
+https://conda.anaconda.org/conda-forge/linux-64/pango-1.50.14-hd33c08f_0.conda#a8b9e35dd7be2c945b0de4fe19a7c3a9
+https://conda.anaconda.org/conda-forge/linux-64/parallelio-2.5.10-mpi_mpich_h773ea27_101.conda#0793f7cf646b9bf66d83d93394474083
+https://conda.anaconda.org/conda-forge/noarch/pyopenssl-23.0.0-pyhd8ed1ab_0.conda#d41957700e83bbb925928764cb7f8878
+https://conda.anaconda.org/conda-forge/noarch/virtualenv-20.20.0-pyhd8ed1ab_0.conda#a4c92707c28aafc95208c747db80fd3f
+https://conda.anaconda.org/conda-forge/linux-64/esmf-8.4.0-mpi_mpich_h7b33e6e_105.conda#14f813a98a4158556c50084bf8e46a78
https://conda.anaconda.org/conda-forge/linux-64/gtk2-2.24.33-h90689f9_2.tar.bz2#957a0255ab58aaf394a91725d73ab422
https://conda.anaconda.org/conda-forge/linux-64/librsvg-2.54.4-h7abd40a_0.tar.bz2#921e53675ed5ea352f022b79abab076a
-https://conda.anaconda.org/conda-forge/linux-64/pre-commit-2.20.0-py310hff52083_1.tar.bz2#8c151d720f9fe3b9962efe71fc10b07b
-https://conda.anaconda.org/conda-forge/linux-64/qt-main-5.15.6-he99da89_3.conda#b7b364a82ad3ce9e56f0bad77efa9ab1
-https://conda.anaconda.org/conda-forge/noarch/urllib3-1.26.13-pyhd8ed1ab_0.conda#3078ef2359efd6ecadbc7e085c5e0592
-https://conda.anaconda.org/conda-forge/linux-64/esmpy-8.2.0-mpi_mpich_py310hd9c82d4_101.tar.bz2#0333d51ee594be40f50b157ac6f27b5a
-https://conda.anaconda.org/conda-forge/linux-64/graphviz-6.0.2-h99bc08f_0.conda#8f247587d1520a2bbc6f79a821b74c07
-https://conda.anaconda.org/conda-forge/linux-64/pyqt-5.15.7-py310h29803b5_2.tar.bz2#1e2c49215b17e6cf06edf100c9869ebe
-https://conda.anaconda.org/conda-forge/noarch/requests-2.28.1-pyhd8ed1ab_1.tar.bz2#089382ee0e2dc2eae33a04cc3c2bddb0
-https://conda.anaconda.org/conda-forge/linux-64/matplotlib-3.6.2-py310hff52083_0.tar.bz2#aa78d12708912cd34135e6694a046ba0
+https://conda.anaconda.org/conda-forge/linux-64/pre-commit-3.1.1-py310hff52083_0.conda#759bfcb929decd0dfa0489070c9cc992
+https://conda.anaconda.org/conda-forge/linux-64/qt-main-5.15.8-h5d23da1_6.conda#59c73debd9405771690ddbbad6c57b69
+https://conda.anaconda.org/conda-forge/noarch/urllib3-1.26.14-pyhd8ed1ab_0.conda#01f33ad2e0aaf6b5ba4add50dad5ad29
+https://conda.anaconda.org/conda-forge/linux-64/esmpy-8.4.0-mpi_mpich_py310h515c5ea_102.conda#bf8276009073388b7159736877eccd79
+https://conda.anaconda.org/conda-forge/linux-64/graphviz-7.1.0-h2e5815a_0.conda#e7ecda996c443142a0e9c379f3b28e48
+https://conda.anaconda.org/conda-forge/linux-64/pyqt-5.15.7-py310hab646b1_3.conda#d049da3204bf5ecb54a852b622f2d7d2
+https://conda.anaconda.org/conda-forge/noarch/requests-2.28.2-pyhd8ed1ab_0.conda#11d178fc55199482ee48d6812ea83983
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-3.7.0-py310hff52083_0.conda#215e2a4504900bef6d68f520c12ef800
+https://conda.anaconda.org/conda-forge/noarch/pooch-1.7.0-pyhd8ed1ab_0.conda#eb2e0fc33ad0d04b51f9280360c13c1e
https://conda.anaconda.org/conda-forge/noarch/sphinx-4.5.0-pyh6c4a22f_0.tar.bz2#46b38d88c4270ff9ba78a89c83c66345
-https://conda.anaconda.org/conda-forge/noarch/pydata-sphinx-theme-0.8.1-pyhd8ed1ab_0.tar.bz2#7d8390ec71225ea9841b276552fdffba
+https://conda.anaconda.org/conda-forge/noarch/pydata-sphinx-theme-0.13.0-pyhd8ed1ab_0.conda#d9916a8dd3f0ee9c795109ee76c5dee6
+https://conda.anaconda.org/conda-forge/linux-64/scipy-1.10.1-py310h8deb116_0.conda#4c9604c5ec179c21f8f0a09e3c164480
https://conda.anaconda.org/conda-forge/noarch/sphinx-copybutton-0.5.0-pyhd8ed1ab_0.tar.bz2#4c969cdd5191306c269490f7ff236d9c
https://conda.anaconda.org/conda-forge/noarch/sphinx-gallery-0.11.1-pyhd8ed1ab_0.tar.bz2#729254314a5d178eefca50acbc2687b8
https://conda.anaconda.org/conda-forge/noarch/sphinx-panels-0.6.0-pyhd8ed1ab_0.tar.bz2#6eec6480601f5d15babf9c3b3987f34a
+https://conda.anaconda.org/conda-forge/linux-64/cartopy-0.21.1-py310hcb7e713_0.conda#bd14eaad9bbf54b78e48ecb8b644fcf6
+https://conda.anaconda.org/conda-forge/noarch/imagehash-4.3.1-pyhd8ed1ab_0.tar.bz2#132ad832787a2156be1f1b309835001a
diff --git a/requirements/ci/nox.lock/py38-linux-64.lock b/requirements/ci/nox.lock/py38-linux-64.lock
index 0836e3d018..b52f0f8d44 100644
--- a/requirements/ci/nox.lock/py38-linux-64.lock
+++ b/requirements/ci/nox.lock/py38-linux-64.lock
@@ -1,6 +1,6 @@
# Generated by conda-lock.
# platform: linux-64
-# input_hash: dc794a12a2155d2a605b34fc34ece8039a0f0d43fbf7d304366cf8c33cf94cd1
+# input_hash: fb647c05bdf2998763af9a184ece4f66796aff1cff2ae207f504c94e6062acaf
@EXPLICIT
https://conda.anaconda.org/conda-forge/linux-64/_libgcc_mutex-0.1-conda_forge.tar.bz2#d7c89558ba9fa0495403155b64376d81
https://conda.anaconda.org/conda-forge/linux-64/ca-certificates-2022.12.7-ha878542_0.conda#ff9f73d45c4a07d6f424495288a26080
@@ -8,7 +8,7 @@ https://conda.anaconda.org/conda-forge/noarch/font-ttf-dejavu-sans-mono-2.37-hab
https://conda.anaconda.org/conda-forge/noarch/font-ttf-inconsolata-3.000-h77eed37_0.tar.bz2#34893075a5c9e55cdafac56607368fc6
https://conda.anaconda.org/conda-forge/noarch/font-ttf-source-code-pro-2.038-h77eed37_0.tar.bz2#4d59c254e01d9cde7957100457e2d5fb
https://conda.anaconda.org/conda-forge/noarch/font-ttf-ubuntu-0.83-hab24e00_0.tar.bz2#19410c3df09dfb12d1206132a1d357c5
-https://conda.anaconda.org/conda-forge/linux-64/ld_impl_linux-64-2.39-hcc3a1bd_1.conda#737be0d34c22d24432049ab7a3214de4
+https://conda.anaconda.org/conda-forge/linux-64/ld_impl_linux-64-2.40-h41732ed_0.conda#7aca3059a1729aa76c597603f10b0dd3
https://conda.anaconda.org/conda-forge/linux-64/libgfortran5-12.2.0-h337968e_19.tar.bz2#164b4b1acaedc47ee7e658ae6b308ca3
https://conda.anaconda.org/conda-forge/linux-64/libstdcxx-ng-12.2.0-h46fd767_19.tar.bz2#1030b1f38c129f2634eae026f704fe60
https://conda.anaconda.org/conda-forge/linux-64/mpi-1.0-mpich.tar.bz2#c1fcff3417b5a22bbc4cf6e8c23648cf
@@ -32,13 +32,14 @@ https://conda.anaconda.org/conda-forge/linux-64/giflib-5.2.1-h36c2ea0_2.tar.bz2#
https://conda.anaconda.org/conda-forge/linux-64/graphite2-1.3.13-h58526e2_1001.tar.bz2#8c54672728e8ec6aa6db90cf2806d220
https://conda.anaconda.org/conda-forge/linux-64/gstreamer-orc-0.4.33-h166bdaf_0.tar.bz2#879c93426c9d0b84a9de4513fbce5f4f
https://conda.anaconda.org/conda-forge/linux-64/icu-70.1-h27087fc_0.tar.bz2#87473a15119779e021c314249d4b4aed
-https://conda.anaconda.org/conda-forge/linux-64/jpeg-9e-h166bdaf_2.tar.bz2#ee8b844357a0946870901c7c6f418268
+https://conda.anaconda.org/conda-forge/linux-64/jpeg-9e-h0b41bf4_3.conda#c7a069243e1fbe9a556ed2ec030e6407
https://conda.anaconda.org/conda-forge/linux-64/keyutils-1.6.1-h166bdaf_0.tar.bz2#30186d27e2c9fa62b45fb1476b7200e3
https://conda.anaconda.org/conda-forge/linux-64/lame-3.100-h166bdaf_1003.tar.bz2#a8832b479f93521a9e7b5b743803be51
https://conda.anaconda.org/conda-forge/linux-64/lerc-4.0.0-h27087fc_0.tar.bz2#76bbff344f0134279f225174e9064c8f
+https://conda.anaconda.org/conda-forge/linux-64/libaec-1.0.6-hcb278e6_1.conda#0f683578378cddb223e7fd24f785ab2a
https://conda.anaconda.org/conda-forge/linux-64/libbrotlicommon-1.0.9-h166bdaf_8.tar.bz2#9194c9bf9428035a05352d031462eae4
https://conda.anaconda.org/conda-forge/linux-64/libdb-6.2.32-h9c3ff4c_0.tar.bz2#3f3258d8f841fbac63b36b75bdac1afd
-https://conda.anaconda.org/conda-forge/linux-64/libdeflate-1.14-h166bdaf_0.tar.bz2#fc84a0446e4e4fb882e78d786cfb9734
+https://conda.anaconda.org/conda-forge/linux-64/libdeflate-1.17-h0b41bf4_0.conda#5cc781fd91968b11a8a7fdbee0982676
https://conda.anaconda.org/conda-forge/linux-64/libev-4.33-h516909a_1.tar.bz2#6f8720dff19e17ce5d48cfe7f3d2f0a3
https://conda.anaconda.org/conda-forge/linux-64/libffi-3.4.2-h7f98852_5.tar.bz2#d645c6d2ac96843a2bfaccd2d62b3ac3
https://conda.anaconda.org/conda-forge/linux-64/libiconv-1.17-h166bdaf_0.tar.bz2#b62b52da46c39ee2bc3c162ac7f1804d
@@ -47,17 +48,17 @@ https://conda.anaconda.org/conda-forge/linux-64/libnsl-2.0.0-h7f98852_0.tar.bz2#
https://conda.anaconda.org/conda-forge/linux-64/libogg-1.3.4-h7f98852_1.tar.bz2#6e8cc2173440d77708196c5b93771680
https://conda.anaconda.org/conda-forge/linux-64/libopenblas-0.3.21-pthreads_h78a6416_3.tar.bz2#8c5963a49b6035c40646a763293fbb35
https://conda.anaconda.org/conda-forge/linux-64/libopus-1.3.1-h7f98852_1.tar.bz2#15345e56d527b330e1cacbdf58676e8f
-https://conda.anaconda.org/conda-forge/linux-64/libtool-2.4.6-h9c3ff4c_1008.tar.bz2#16e143a1ed4b4fd169536373957f6fee
-https://conda.anaconda.org/conda-forge/linux-64/libudev1-252-h166bdaf_0.tar.bz2#174243089ec111479298a5b7099b64b5
+https://conda.anaconda.org/conda-forge/linux-64/libtool-2.4.7-h27087fc_0.conda#f204c8ba400ec475452737094fb81d52
+https://conda.anaconda.org/conda-forge/linux-64/libudev1-253-h0b41bf4_0.conda#6c2addbd9aa4ee47c76d50c9f0df8cd6
https://conda.anaconda.org/conda-forge/linux-64/libuuid-2.32.1-h7f98852_1000.tar.bz2#772d69f030955d9646d3d0eaf21d859d
https://conda.anaconda.org/conda-forge/linux-64/libwebp-base-1.2.4-h166bdaf_0.tar.bz2#ac2ccf7323d21f2994e4d1f5da664f37
https://conda.anaconda.org/conda-forge/linux-64/libzlib-1.2.13-h166bdaf_4.tar.bz2#f3f9de449d32ca9b9c66a22863c96f41
-https://conda.anaconda.org/conda-forge/linux-64/lz4-c-1.9.3-h9c3ff4c_1.tar.bz2#fbe97e8fa6f275d7c76a09e795adc3e6
-https://conda.anaconda.org/conda-forge/linux-64/mpg123-1.31.1-h27087fc_0.tar.bz2#0af513b75f78a701a152568a31303bdf
+https://conda.anaconda.org/conda-forge/linux-64/lz4-c-1.9.4-hcb278e6_0.conda#318b08df404f9c9be5712aaa5a6f0bb0
+https://conda.anaconda.org/conda-forge/linux-64/mpg123-1.31.2-hcb278e6_0.conda#08efb1e1813f1a151b7a945b972a049b
https://conda.anaconda.org/conda-forge/linux-64/mpich-4.0.3-h846660c_100.tar.bz2#50d66bb751cfa71ee2a48b2d3eb90ac1
https://conda.anaconda.org/conda-forge/linux-64/ncurses-6.3-h27087fc_1.tar.bz2#4acfc691e64342b9dae57cf2adc63238
https://conda.anaconda.org/conda-forge/linux-64/nspr-4.35-h27087fc_0.conda#da0ec11a6454ae19bff5b02ed881a2b1
-https://conda.anaconda.org/conda-forge/linux-64/openssl-3.0.7-h0b41bf4_1.conda#7adaac6ff98219bcb99b45e408b80f4e
+https://conda.anaconda.org/conda-forge/linux-64/openssl-3.0.8-h0b41bf4_0.conda#e043403cd18faf815bf7705ab6c1e092
https://conda.anaconda.org/conda-forge/linux-64/pixman-0.40.0-h36c2ea0_0.tar.bz2#660e72c82f2e75a6b3fe6a6e75c79f19
https://conda.anaconda.org/conda-forge/linux-64/pthread-stubs-0.4-h36c2ea0_1001.tar.bz2#22dad4df6e8630e8dff2428f6f6a7036
https://conda.anaconda.org/conda-forge/linux-64/xorg-kbproto-1.0.7-h7f98852_1002.tar.bz2#4b230e8381279d76131116660f5a241a
@@ -65,12 +66,12 @@ https://conda.anaconda.org/conda-forge/linux-64/xorg-libice-1.0.10-h7f98852_0.ta
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxau-1.0.9-h7f98852_0.tar.bz2#bf6f803a544f26ebbdc3bfff272eb179
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxdmcp-1.1.3-h7f98852_0.tar.bz2#be93aabceefa2fac576e971aef407908
https://conda.anaconda.org/conda-forge/linux-64/xorg-renderproto-0.11.1-h7f98852_1002.tar.bz2#06feff3d2634e3097ce2fe681474b534
-https://conda.anaconda.org/conda-forge/linux-64/xorg-xextproto-7.3.0-h7f98852_1002.tar.bz2#1e15f6ad85a7d743a2ac68dae6c82b98
+https://conda.anaconda.org/conda-forge/linux-64/xorg-xextproto-7.3.0-h0b41bf4_1003.conda#bce9f945da8ad2ae9b1d7165a64d0f87
https://conda.anaconda.org/conda-forge/linux-64/xorg-xproto-7.0.31-h7f98852_1007.tar.bz2#b4a4381d54784606820704f7b5f05a15
-https://conda.anaconda.org/conda-forge/linux-64/xxhash-0.8.0-h7f98852_3.tar.bz2#52402c791f35e414e704b7a113f99605
+https://conda.anaconda.org/conda-forge/linux-64/xxhash-0.8.1-h0b41bf4_0.conda#e9c3bcf0e0c719431abec8ca447eee27
https://conda.anaconda.org/conda-forge/linux-64/xz-5.2.6-h166bdaf_0.tar.bz2#2161070d867d1b1204ea749c8eec4ef0
https://conda.anaconda.org/conda-forge/linux-64/yaml-0.2.5-h7f98852_2.tar.bz2#4cb3ad778ec2d5a7acbdf254eb1c42ae
-https://conda.anaconda.org/conda-forge/linux-64/jack-1.9.21-h583fa2b_2.conda#7b36a10b58964d4444fcba44244710c5
+https://conda.anaconda.org/conda-forge/linux-64/jack-1.9.22-h11f4161_0.conda#504fa9e712b99494a9cf4630e3ca7d78
https://conda.anaconda.org/conda-forge/linux-64/libblas-3.9.0-16_linux64_openblas.tar.bz2#d9b7a8639171f6c6fa0a983edabcfe2b
https://conda.anaconda.org/conda-forge/linux-64/libbrotlidec-1.0.9-h166bdaf_8.tar.bz2#4ae4d7795d33e02bd20f6b23d91caf82
https://conda.anaconda.org/conda-forge/linux-64/libbrotlienc-1.0.9-h166bdaf_8.tar.bz2#04bac51ba35ea023dc48af73c1c88c25
@@ -78,8 +79,8 @@ https://conda.anaconda.org/conda-forge/linux-64/libcap-2.66-ha37c62d_0.tar.bz2#2
https://conda.anaconda.org/conda-forge/linux-64/libedit-3.1.20191231-he28a2e2_2.tar.bz2#4d331e44109e3f0e19b4cb8f9b82f3e1
https://conda.anaconda.org/conda-forge/linux-64/libevent-2.1.10-h28343ad_4.tar.bz2#4a049fc560e00e43151dc51368915fdd
https://conda.anaconda.org/conda-forge/linux-64/libflac-1.4.2-h27087fc_0.tar.bz2#7daf72d8e2a8e848e11d63ed6d1026e0
-https://conda.anaconda.org/conda-forge/linux-64/libgpg-error-1.45-hc0c96e0_0.tar.bz2#839aeb24ab885a7b902247a6d943d02f
-https://conda.anaconda.org/conda-forge/linux-64/libnghttp2-1.47.0-hff17c54_1.tar.bz2#2b7dbfa6988a41f9d23ba6d4f0e1d74e
+https://conda.anaconda.org/conda-forge/linux-64/libgpg-error-1.46-h620e276_0.conda#27e745f6f2e4b757e95dd7225fbe6bdb
+https://conda.anaconda.org/conda-forge/linux-64/libnghttp2-1.52.0-h61bc06f_0.conda#613955a50485812985c059e7b269f42e
https://conda.anaconda.org/conda-forge/linux-64/libpng-1.6.39-h753d276_0.conda#e1c890aebdebbfbf87e2c917187b4416
https://conda.anaconda.org/conda-forge/linux-64/libsqlite-3.40.0-h753d276_0.tar.bz2#2e5f9a37d487e1019fd4d8113adb2f9f
https://conda.anaconda.org/conda-forge/linux-64/libssh2-1.10.0-hf14f497_3.tar.bz2#d85acad4b47dff4e3def14a769a97906
@@ -87,78 +88,77 @@ https://conda.anaconda.org/conda-forge/linux-64/libvorbis-1.3.7-h9c3ff4c_0.tar.b
https://conda.anaconda.org/conda-forge/linux-64/libxcb-1.13-h7f98852_1004.tar.bz2#b3653fdc58d03face9724f602218a904
https://conda.anaconda.org/conda-forge/linux-64/libxml2-2.10.3-h7463322_0.tar.bz2#3b933ea47ef8f330c4c068af25fcd6a8
https://conda.anaconda.org/conda-forge/linux-64/libzip-1.9.2-hc929e4a_1.tar.bz2#5b122b50e738c4be5c3f2899f010d7cf
-https://conda.anaconda.org/conda-forge/linux-64/mysql-common-8.0.31-h26416b9_0.tar.bz2#6c531bc30d49ae75b9c7c7f65bd62e3c
+https://conda.anaconda.org/conda-forge/linux-64/mysql-common-8.0.32-ha901b37_0.conda#6a39818710235826181e104aada40c75
https://conda.anaconda.org/conda-forge/linux-64/pcre2-10.40-hc3806b6_0.tar.bz2#69e2c796349cd9b273890bee0febfe1b
https://conda.anaconda.org/conda-forge/linux-64/readline-8.1.2-h0f457ee_0.tar.bz2#db2ebbe2943aae81ed051a6a9af8e0fa
https://conda.anaconda.org/conda-forge/linux-64/tk-8.6.12-h27826a3_0.tar.bz2#5b8c42eb62e9fc961af70bdd6a26e168
https://conda.anaconda.org/conda-forge/linux-64/udunits2-2.2.28-hc3e0081_0.tar.bz2#d4c341e0379c31e9e781d4f204726867
https://conda.anaconda.org/conda-forge/linux-64/xorg-libsm-1.2.3-hd9c2040_1000.tar.bz2#9e856f78d5c80d5a78f61e72d1d473a3
https://conda.anaconda.org/conda-forge/linux-64/zlib-1.2.13-h166bdaf_4.tar.bz2#4b11e365c0275b808be78b30f904e295
-https://conda.anaconda.org/conda-forge/linux-64/zstd-1.5.2-h6239696_4.tar.bz2#adcf0be7897e73e312bd24353b613f74
+https://conda.anaconda.org/conda-forge/linux-64/zstd-1.5.2-h3eb15da_6.conda#6b63daed8feeca47be78f323e793d555
https://conda.anaconda.org/conda-forge/linux-64/brotli-bin-1.0.9-h166bdaf_8.tar.bz2#e5613f2bc717e9945840ff474419b8e4
https://conda.anaconda.org/conda-forge/linux-64/freetype-2.12.1-hca18f0e_1.conda#e1232042de76d24539a436d37597eb06
https://conda.anaconda.org/conda-forge/linux-64/hdf4-4.2.15-h9772cbc_5.tar.bz2#ee08782aff2ff9b3291c967fa6bc7336
-https://conda.anaconda.org/conda-forge/linux-64/krb5-1.19.3-h08a2579_0.tar.bz2#d25e05e7ee0e302b52d24491db4891eb
+https://conda.anaconda.org/conda-forge/linux-64/krb5-1.20.1-h81ceb04_0.conda#89a41adce7106749573d883b2f657d78
https://conda.anaconda.org/conda-forge/linux-64/libcblas-3.9.0-16_linux64_openblas.tar.bz2#20bae26d0a1db73f758fc3754cab4719
https://conda.anaconda.org/conda-forge/linux-64/libgcrypt-1.10.1-h166bdaf_0.tar.bz2#f967fc95089cd247ceed56eda31de3a9
https://conda.anaconda.org/conda-forge/linux-64/libglib-2.74.1-h606061b_1.tar.bz2#ed5349aa96776e00b34eccecf4a948fe
https://conda.anaconda.org/conda-forge/linux-64/liblapack-3.9.0-16_linux64_openblas.tar.bz2#955d993f41f9354bf753d29864ea20ad
-https://conda.anaconda.org/conda-forge/linux-64/libllvm15-15.0.6-h63197d8_0.conda#201168ef66095bbd565e124ee2c56a20
-https://conda.anaconda.org/conda-forge/linux-64/libsndfile-1.1.0-hcb278e6_1.conda#d7a07b1f5974bce4735112aaef0c1467
-https://conda.anaconda.org/conda-forge/linux-64/libtiff-4.4.0-h55922b4_4.tar.bz2#901791f0ec7cddc8714e76e273013a91
-https://conda.anaconda.org/conda-forge/linux-64/libxkbcommon-1.0.3-he3ba5ed_0.tar.bz2#f9dbabc7e01c459ed7a1d1d64b206e9b
-https://conda.anaconda.org/conda-forge/linux-64/mysql-libs-8.0.31-hbc51c84_0.tar.bz2#da9633eee814d4e910fe42643a356315
-https://conda.anaconda.org/conda-forge/linux-64/nss-3.82-he02c5a1_0.conda#f8d7f11d19e4cb2207eab159fd4c0152
-https://conda.anaconda.org/conda-forge/linux-64/python-3.8.15-h4a9ceb5_0_cpython.conda#dc29a8a79d0f2c80004cc06d3190104f
+https://conda.anaconda.org/conda-forge/linux-64/libllvm15-15.0.7-hadd5161_0.conda#70cbb0c2033665f2a7339bf0ec51a67f
+https://conda.anaconda.org/conda-forge/linux-64/libsndfile-1.2.0-hb75c966_0.conda#c648d19cd9c8625898d5d370414de7c7
+https://conda.anaconda.org/conda-forge/linux-64/libtiff-4.5.0-h6adf6a1_2.conda#2e648a34072eb39d7c4fc2a9981c5f0c
+https://conda.anaconda.org/conda-forge/linux-64/libxkbcommon-1.5.0-h79f4944_0.conda#3f67368c9b0e77a693acad193310baf1
+https://conda.anaconda.org/conda-forge/linux-64/mysql-libs-8.0.32-hd7da12d_0.conda#b05d7ea8b76f1172d5fe4f30e03277ea
+https://conda.anaconda.org/conda-forge/linux-64/nss-3.88-he45b914_0.conda#d7a81dfb99ad8fbb88872fb7ec646e6c
+https://conda.anaconda.org/conda-forge/linux-64/python-3.8.16-he550d4f_1_cpython.conda#9de84cccfbc5f8350a3667bb6ef6fc30
https://conda.anaconda.org/conda-forge/linux-64/sqlite-3.40.0-h4ff8645_0.tar.bz2#bb11803129cbbb53ed56f9506ff74145
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-0.4.0-h166bdaf_0.tar.bz2#384e7fcb3cd162ba3e4aed4b687df566
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-keysyms-0.4.0-h166bdaf_0.tar.bz2#637054603bb7594302e3bf83f0a99879
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-renderutil-0.3.9-h166bdaf_0.tar.bz2#732e22f1741bccea861f5668cf7342a7
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-wm-0.4.1-h166bdaf_0.tar.bz2#0a8e20a8aef954390b9481a527421a8c
-https://conda.anaconda.org/conda-forge/linux-64/xorg-libx11-1.7.2-h7f98852_0.tar.bz2#12a61e640b8894504326aadafccbb790
-https://conda.anaconda.org/conda-forge/noarch/alabaster-0.7.12-py_0.tar.bz2#2489a97287f90176ecdc3ca982b4b0a0
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libx11-1.8.4-h0b41bf4_0.conda#ea8fbfeb976ac49cbeb594e985393514
+https://conda.anaconda.org/conda-forge/noarch/alabaster-0.7.13-pyhd8ed1ab_0.conda#06006184e203b61d3525f90de394471e
https://conda.anaconda.org/conda-forge/linux-64/antlr-python-runtime-4.7.2-py38h578d9bd_1003.tar.bz2#db8b471d9a764f561a129f94ea215c0a
https://conda.anaconda.org/conda-forge/linux-64/atk-1.0-2.38.0-hd4edc92_1.tar.bz2#6c72ec3e660a51736913ef6ea68c454b
-https://conda.anaconda.org/conda-forge/noarch/attrs-22.1.0-pyh71513ae_1.tar.bz2#6d3ccbc56256204925bfa8378722792f
+https://conda.anaconda.org/conda-forge/noarch/attrs-22.2.0-pyh71513ae_0.conda#8b76db7818a4e401ed4486c4c1635cd9
https://conda.anaconda.org/conda-forge/linux-64/brotli-1.0.9-h166bdaf_8.tar.bz2#2ff08978892a3e8b954397c461f18418
https://conda.anaconda.org/conda-forge/noarch/certifi-2022.12.7-pyhd8ed1ab_0.conda#fb9addc3db06e56abe03e0e9f21a63e6
https://conda.anaconda.org/conda-forge/noarch/cfgv-3.3.1-pyhd8ed1ab_0.tar.bz2#ebb5f5f7dc4f1a3780ef7ea7738db08c
https://conda.anaconda.org/conda-forge/noarch/charset-normalizer-2.1.1-pyhd8ed1ab_0.tar.bz2#c1d5b294fbf9a795dec349a6f4d8be8e
https://conda.anaconda.org/conda-forge/noarch/click-8.1.3-unix_pyhd8ed1ab_2.tar.bz2#20e4087407c7cb04a40817114b333dbf
-https://conda.anaconda.org/conda-forge/noarch/cloudpickle-2.2.0-pyhd8ed1ab_0.tar.bz2#a6cf47b09786423200d7982d1faa19eb
+https://conda.anaconda.org/conda-forge/noarch/cloudpickle-2.2.1-pyhd8ed1ab_0.conda#b325bfc4cff7d7f8a868f1f7ecc4ed16
https://conda.anaconda.org/conda-forge/noarch/colorama-0.4.6-pyhd8ed1ab_0.tar.bz2#3faab06a954c2a04039983f2c4a50d99
https://conda.anaconda.org/conda-forge/noarch/cycler-0.11.0-pyhd8ed1ab_0.tar.bz2#a50559fad0affdbb33729a68669ca1cb
https://conda.anaconda.org/conda-forge/linux-64/dbus-1.13.6-h5008d03_3.tar.bz2#ecfff944ba3960ecb334b9a2663d708d
https://conda.anaconda.org/conda-forge/noarch/distlib-0.3.6-pyhd8ed1ab_0.tar.bz2#b65b4d50dbd2d50fa0aeac367ec9eed7
https://conda.anaconda.org/conda-forge/linux-64/docutils-0.17.1-py38h578d9bd_3.tar.bz2#34e1f12e3ed15aff218644e9d865b722
-https://conda.anaconda.org/conda-forge/noarch/exceptiongroup-1.0.4-pyhd8ed1ab_0.tar.bz2#e0734d1f12de77f9daca98bda3428733
+https://conda.anaconda.org/conda-forge/noarch/exceptiongroup-1.1.0-pyhd8ed1ab_0.conda#a385c3e8968b4cf8fbc426ace915fd1a
https://conda.anaconda.org/conda-forge/noarch/execnet-1.9.0-pyhd8ed1ab_0.tar.bz2#0e521f7a5e60d508b121d38b04874fb2
-https://conda.anaconda.org/conda-forge/noarch/filelock-3.8.2-pyhd8ed1ab_0.conda#0f09c2bc17ddd8732be8e5b99297c7ce
-https://conda.anaconda.org/conda-forge/linux-64/fontconfig-2.14.1-hc2a2eb6_0.tar.bz2#78415f0180a8d9c5bcc47889e00d5fb1
-https://conda.anaconda.org/conda-forge/noarch/fsspec-2022.11.0-pyhd8ed1ab_0.tar.bz2#eb919f2119a6db5d0192f9e9c3711572
-https://conda.anaconda.org/conda-forge/linux-64/gdk-pixbuf-2.42.8-hff1cb4f_1.tar.bz2#a61c6312192e7c9de71548a6706a21e6
+https://conda.anaconda.org/conda-forge/noarch/filelock-3.9.0-pyhd8ed1ab_0.conda#1addc115923d646ca19ed90edc413506
+https://conda.anaconda.org/conda-forge/linux-64/fontconfig-2.14.2-h14ed4e7_0.conda#0f69b688f52ff6da70bccb7ff7001d1d
+https://conda.anaconda.org/conda-forge/noarch/fsspec-2023.1.0-pyhd8ed1ab_0.conda#44f6828b8f7cc3433d68d1d1c0e9add2
+https://conda.anaconda.org/conda-forge/linux-64/gdk-pixbuf-2.42.10-h05c8ddd_0.conda#1a109126a43003d65b39c1cad656bc9b
https://conda.anaconda.org/conda-forge/linux-64/glib-tools-2.74.1-h6239696_1.tar.bz2#5f442e6bc9d89ba236eb25a25c5c2815
https://conda.anaconda.org/conda-forge/linux-64/gts-0.7.6-h64030ff_2.tar.bz2#112eb9b5b93f0c02e59aea4fd1967363
https://conda.anaconda.org/conda-forge/noarch/idna-3.4-pyhd8ed1ab_0.tar.bz2#34272b248891bddccc64479f9a7fffed
https://conda.anaconda.org/conda-forge/noarch/imagesize-1.4.1-pyhd8ed1ab_0.tar.bz2#7de5386c8fea29e76b303f37dde4c352
-https://conda.anaconda.org/conda-forge/noarch/iniconfig-1.1.1-pyh9f0ad1d_0.tar.bz2#39161f81cc5e5ca45b8226fbb06c6905
+https://conda.anaconda.org/conda-forge/noarch/iniconfig-2.0.0-pyhd8ed1ab_0.conda#f800d2da156d08e289b14e87e43c1ae5
https://conda.anaconda.org/conda-forge/noarch/iris-sample-data-2.4.0-pyhd8ed1ab_0.tar.bz2#18ee9c07cf945a33f92caf1ee3d23ad9
https://conda.anaconda.org/conda-forge/linux-64/kiwisolver-1.4.4-py38h43d8883_1.tar.bz2#41ca56d5cac7bfc7eb4fcdbee878eb84
-https://conda.anaconda.org/conda-forge/linux-64/lcms2-2.14-h6ed2654_0.tar.bz2#dcc588839de1445d90995a0a2c4f3a39
-https://conda.anaconda.org/conda-forge/linux-64/libclang13-15.0.6-default_h3a83d3e_0.conda#535dd0ca1dcb165b6a8ffa10d01945fe
-https://conda.anaconda.org/conda-forge/linux-64/libcups-2.3.3-h3e49a29_2.tar.bz2#3b88f1d0fe2580594d58d7e44d664617
-https://conda.anaconda.org/conda-forge/linux-64/libcurl-7.86.0-h2283fc2_1.tar.bz2#fdca8cd67ec2676f90a70ac73a32538b
-https://conda.anaconda.org/conda-forge/linux-64/libpq-15.1-h67c24c5_1.conda#e1389a8d9a907133b3e6483c2807d243
+https://conda.anaconda.org/conda-forge/linux-64/lcms2-2.14-hfd0df8a_1.conda#c2566c2ea5f153ddd6bf4acaf7547d97
+https://conda.anaconda.org/conda-forge/linux-64/libclang13-15.0.7-default_h3e3d535_1.conda#a3a0f7a6f0885f5e1e0ec691566afb77
+https://conda.anaconda.org/conda-forge/linux-64/libcups-2.3.3-h36d4200_3.conda#c9f4416a34bc91e0eb029f912c68f81f
+https://conda.anaconda.org/conda-forge/linux-64/libcurl-7.88.1-hdc1c0ab_0.conda#81eaeb3b35163c8e90e57532bc93754d
+https://conda.anaconda.org/conda-forge/linux-64/libpq-15.2-hb675445_0.conda#4654b17eccaba55b8581d6b9c77f53cc
https://conda.anaconda.org/conda-forge/linux-64/libsystemd0-252-h2a991cd_0.tar.bz2#3c5ae9f61f663b3d5e1bf7f7da0c85f5
-https://conda.anaconda.org/conda-forge/linux-64/libwebp-1.2.4-h522a892_0.tar.bz2#802e43f480122a85ae6a34c1909f8f98
+https://conda.anaconda.org/conda-forge/linux-64/libwebp-1.2.4-h1daa5a0_1.conda#77003f63d1763c1e6569a02c1742c9f4
https://conda.anaconda.org/conda-forge/noarch/locket-1.0.0-pyhd8ed1ab_0.tar.bz2#91e27ef3d05cc772ce627e51cff111c4
-https://conda.anaconda.org/conda-forge/linux-64/markupsafe-2.1.1-py38h0a891b7_2.tar.bz2#c342a370480791db83d5dd20f2d8899f
+https://conda.anaconda.org/conda-forge/linux-64/markupsafe-2.1.2-py38h1de0b5d_0.conda#6d97b5d6f06933ab653f1862ddf6e33e
https://conda.anaconda.org/conda-forge/linux-64/mpi4py-3.1.4-py38h97ac3a3_0.tar.bz2#0c469687a517052c0d581fc6e1a4189d
https://conda.anaconda.org/conda-forge/noarch/munkres-1.1.4-pyh9f0ad1d_0.tar.bz2#2ba8498c1018c1e9c61eb99b973dfe19
-https://conda.anaconda.org/conda-forge/linux-64/numpy-1.23.5-py38h7042d01_0.conda#d5a3620cd8c1af4115120f21d678507a
-https://conda.anaconda.org/conda-forge/linux-64/openjpeg-2.5.0-h7d73246_1.tar.bz2#a11b4df9271a8d7917686725aa04c8f2
-https://conda.anaconda.org/conda-forge/noarch/packaging-22.0-pyhd8ed1ab_0.conda#0e8e1bd93998978fc3125522266d12db
-https://conda.anaconda.org/conda-forge/noarch/platformdirs-2.6.0-pyhd8ed1ab_0.conda#b1b2ab02d1ece1719f7fa002ad4bc70d
+https://conda.anaconda.org/conda-forge/linux-64/numpy-1.24.2-py38h10c12cc_0.conda#05592c85b9f6931dc2df1e80c0d56294
+https://conda.anaconda.org/conda-forge/linux-64/openjpeg-2.5.0-hfec8fc6_2.conda#5ce6a42505c6e9e6151c54c3ec8d68ea
+https://conda.anaconda.org/conda-forge/noarch/packaging-23.0-pyhd8ed1ab_0.conda#1ff2e3ca41f0ce16afec7190db28288b
https://conda.anaconda.org/conda-forge/noarch/pluggy-1.0.0-pyhd8ed1ab_5.tar.bz2#7d301a0d25f424d96175f810935f0da9
https://conda.anaconda.org/conda-forge/noarch/ply-3.11-py_1.tar.bz2#7205635cd71531943440fbfe3b6b5727
https://conda.anaconda.org/conda-forge/linux-64/psutil-5.9.4-py38h0a891b7_0.tar.bz2#fe2ef279417faa1af0adf178de2032f7
@@ -166,16 +166,16 @@ https://conda.anaconda.org/conda-forge/noarch/pycparser-2.21-pyhd8ed1ab_0.tar.bz
https://conda.anaconda.org/conda-forge/noarch/pyparsing-3.0.9-pyhd8ed1ab_0.tar.bz2#e8fbc1b54b25f4b08281467bc13b70cc
https://conda.anaconda.org/conda-forge/noarch/pyshp-2.3.1-pyhd8ed1ab_0.tar.bz2#92a889dc236a5197612bc85bee6d7174
https://conda.anaconda.org/conda-forge/noarch/pysocks-1.7.1-pyha2e5f31_6.tar.bz2#2a7de29fb590ca14b5243c4c812c8025
-https://conda.anaconda.org/conda-forge/linux-64/python-xxhash-3.0.0-py38h0a891b7_2.tar.bz2#9b13816a39904084556126a6ce7fd0d0
-https://conda.anaconda.org/conda-forge/noarch/pytz-2022.6-pyhd8ed1ab_0.tar.bz2#b1f26ad83328e486910ef7f6e81dc061
+https://conda.anaconda.org/conda-forge/linux-64/python-xxhash-3.2.0-py38h1de0b5d_0.conda#7db73572d4f7e10a759bad609a228ad0
+https://conda.anaconda.org/conda-forge/noarch/pytz-2022.7.1-pyhd8ed1ab_0.conda#f59d49a7b464901cf714b9e7984d01a2
https://conda.anaconda.org/conda-forge/linux-64/pyyaml-6.0-py38h0a891b7_5.tar.bz2#0856c59f9ddb710c640dc0428d66b1b7
-https://conda.anaconda.org/conda-forge/noarch/setuptools-65.5.1-pyhd8ed1ab_0.tar.bz2#cfb8dc4d9d285ca5fb1177b9dd450e33
+https://conda.anaconda.org/conda-forge/noarch/setuptools-67.4.0-pyhd8ed1ab_0.conda#c6f4b87020c72e2700e3e94c1fc93b70
https://conda.anaconda.org/conda-forge/noarch/six-1.16.0-pyh6c4a22f_0.tar.bz2#e5f25f8dbc060e9a8d912e432202afc2
https://conda.anaconda.org/conda-forge/noarch/snowballstemmer-2.2.0-pyhd8ed1ab_0.tar.bz2#4d22a9315e78c6827f806065957d566e
https://conda.anaconda.org/conda-forge/noarch/soupsieve-2.3.2.post1-pyhd8ed1ab_0.tar.bz2#146f4541d643d48fc8a75cacf69f03ae
-https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-applehelp-1.0.2-py_0.tar.bz2#20b2eaeaeea4ef9a9a0d99770620fd09
+https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-applehelp-1.0.4-pyhd8ed1ab_0.conda#5a31a7d564f551d0e6dff52fd8cb5b16
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-devhelp-1.0.2-py_0.tar.bz2#68e01cac9d38d0e717cd5c87bc3d2cc9
-https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-htmlhelp-2.0.0-pyhd8ed1ab_0.tar.bz2#77dad82eb9c8c1525ff7953e0756d708
+https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-htmlhelp-2.0.1-pyhd8ed1ab_0.conda#6c8c4d6eb2325e59290ac6dbbeacd5f0
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-jsmath-1.0.1-py_0.tar.bz2#67cd9d9c0382d37479b4d306c369a2d4
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-qthelp-1.0.3-py_0.tar.bz2#d01180388e6d1838c3e1ad029590aa7a
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-serializinghtml-1.1.5-pyhd8ed1ab_2.tar.bz2#9ff55a0901cf952f05c654394de76bf7
@@ -187,78 +187,84 @@ https://conda.anaconda.org/conda-forge/noarch/typing_extensions-4.4.0-pyha770c72
https://conda.anaconda.org/conda-forge/linux-64/unicodedata2-15.0.0-py38h0a891b7_0.tar.bz2#44421904760e9f5ae2035193e04360f0
https://conda.anaconda.org/conda-forge/noarch/wheel-0.38.4-pyhd8ed1ab_0.tar.bz2#c829cfb8cb826acb9de0ac1a2df0a940
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-image-0.4.0-h166bdaf_0.tar.bz2#c9b568bd804cb2903c6be6f5f68182e4
-https://conda.anaconda.org/conda-forge/linux-64/xorg-libxext-1.3.4-h7f98852_1.tar.bz2#536cc5db4d0a3ba0630541aec064b5e4
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxext-1.3.4-h0b41bf4_2.conda#82b6df12252e6f32402b96dacc656fec
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxrender-0.9.10-h7f98852_1003.tar.bz2#f59c1242cc1dd93e72c2ee2b360979eb
-https://conda.anaconda.org/conda-forge/noarch/zipp-3.11.0-pyhd8ed1ab_0.conda#09b5b885341697137879a4f039a9e5a1
-https://conda.anaconda.org/conda-forge/noarch/babel-2.11.0-pyhd8ed1ab_0.tar.bz2#2ea70fde8d581ba9425a761609eed6ba
-https://conda.anaconda.org/conda-forge/noarch/beautifulsoup4-4.11.1-pyha770c72_0.tar.bz2#eeec8814bd97b2681f708bb127478d7d
+https://conda.anaconda.org/conda-forge/noarch/zipp-3.15.0-pyhd8ed1ab_0.conda#13018819ca8f5b7cc675a8faf1f5fedf
+https://conda.anaconda.org/conda-forge/noarch/babel-2.12.1-pyhd8ed1ab_1.conda#ac432e732804a81ddcf29c92ead57cde
+https://conda.anaconda.org/conda-forge/noarch/beautifulsoup4-4.11.2-pyha770c72_0.conda#88b59f6989f0ed5ab3433af0b82555e1
https://conda.anaconda.org/conda-forge/linux-64/cairo-1.16.0-ha61ee94_1014.tar.bz2#d1a88f3ed5b52e1024b80d4bcd26a7a0
-https://conda.anaconda.org/conda-forge/linux-64/cffi-1.15.1-py38h4a40e3a_2.tar.bz2#2276b1f4d1ede3f5f14cc7e4ae6f9a33
+https://conda.anaconda.org/conda-forge/linux-64/cffi-1.15.1-py38h4a40e3a_3.conda#3ac112151c6b6cfe457e976de41af0c5
https://conda.anaconda.org/conda-forge/linux-64/cftime-1.6.2-py38h26c90d9_1.tar.bz2#dcc025a7bb54374979c500c2e161fac9
-https://conda.anaconda.org/conda-forge/linux-64/contourpy-1.0.6-py38h43d8883_0.tar.bz2#1107ee053d55172b26c4fc905dd0238e
-https://conda.anaconda.org/conda-forge/linux-64/curl-7.86.0-h2283fc2_1.tar.bz2#9d4149760567cb232691cce2d8ccc21f
+https://conda.anaconda.org/conda-forge/linux-64/contourpy-1.0.7-py38hfbd4bf9_0.conda#638537863b298151635c05c762a997ab
+https://conda.anaconda.org/conda-forge/linux-64/curl-7.88.1-hdc1c0ab_0.conda#1968e4fef727858ac04746560e820928
https://conda.anaconda.org/conda-forge/linux-64/fonttools-4.38.0-py38h0a891b7_1.tar.bz2#62c89ddefed9c5835e228a32b357a28d
https://conda.anaconda.org/conda-forge/linux-64/glib-2.74.1-h6239696_1.tar.bz2#f3220a9e9d3abcbfca43419a219df7e4
-https://conda.anaconda.org/conda-forge/linux-64/hdf5-1.12.2-mpi_mpich_h5d83325_0.tar.bz2#6b5c2d276f306df759cfbdb0f41c4db9
-https://conda.anaconda.org/conda-forge/noarch/importlib-metadata-5.1.0-pyha770c72_0.conda#46a62e35b9ae515cf0e49afc7fe0e7ef
+https://conda.anaconda.org/conda-forge/linux-64/hdf5-1.12.2-mpi_mpich_h5d83325_1.conda#811c4d55cf17b42336ffa314239717b0
+https://conda.anaconda.org/conda-forge/noarch/importlib-metadata-6.0.0-pyha770c72_0.conda#691644becbcdca9f73243450b1c63e62
+https://conda.anaconda.org/conda-forge/noarch/importlib_resources-5.12.0-pyhd8ed1ab_0.conda#e5fd2260a231ee63b6969f4801082f2b
https://conda.anaconda.org/conda-forge/noarch/jinja2-3.1.2-pyhd8ed1ab_1.tar.bz2#c8490ed5c70966d232fdd389d0dbed37
-https://conda.anaconda.org/conda-forge/linux-64/libclang-15.0.6-default_h2e3cab8_0.conda#1b2cee49acc5b03c73ad0f68bfe04bb8
-https://conda.anaconda.org/conda-forge/linux-64/libgd-2.3.3-h18fbbfe_3.tar.bz2#ea9758cf553476ddf75c789fdd239dc5
+https://conda.anaconda.org/conda-forge/linux-64/libclang-15.0.7-default_had23c3d_1.conda#36c65ed73b7c92589bd9562ef8a6023d
+https://conda.anaconda.org/conda-forge/linux-64/libgd-2.3.3-h5aea950_4.conda#82ef57611ace65b59db35a9687264572
https://conda.anaconda.org/conda-forge/linux-64/mo_pack-0.2.0-py38h26c90d9_1008.tar.bz2#6bc8cd29312f4fc77156b78124e165cd
https://conda.anaconda.org/conda-forge/noarch/nodeenv-1.7.0-pyhd8ed1ab_0.tar.bz2#fbe1182f650c04513046d6894046cd6c
https://conda.anaconda.org/conda-forge/noarch/partd-1.3.0-pyhd8ed1ab_0.tar.bz2#af8c82d121e63082926062d61d9abb54
-https://conda.anaconda.org/conda-forge/linux-64/pillow-9.2.0-py38h9eb91d8_3.tar.bz2#61dc7b3140b7b79b1985b53d52726d74
-https://conda.anaconda.org/conda-forge/noarch/pip-22.3.1-pyhd8ed1ab_0.tar.bz2#da66f2851b9836d3a7c5190082a45f7d
+https://conda.anaconda.org/conda-forge/linux-64/pillow-9.4.0-py38hde6dc18_1.conda#3de5619d3f556f966189e5251a266125
+https://conda.anaconda.org/conda-forge/noarch/pip-23.0.1-pyhd8ed1ab_0.conda#8025ca83b8ba5430b640b83917c2a6f7
https://conda.anaconda.org/conda-forge/noarch/pockets-0.9.1-py_0.tar.bz2#1b52f0c42e8077e5a33e00fe72269364
-https://conda.anaconda.org/conda-forge/linux-64/proj-9.1.0-h93bde94_0.tar.bz2#255c7204dda39747c3ba380d28b026d7
-https://conda.anaconda.org/conda-forge/linux-64/pulseaudio-16.1-h126f2b6_0.tar.bz2#e4b74b33e13dd146e7d8b5078fc9ad30
-https://conda.anaconda.org/conda-forge/noarch/pygments-2.13.0-pyhd8ed1ab_0.tar.bz2#9f478e8eedd301008b5f395bad0caaed
-https://conda.anaconda.org/conda-forge/noarch/pytest-7.2.0-pyhd8ed1ab_2.tar.bz2#ac82c7aebc282e6ac0450fca012ca78c
+https://conda.anaconda.org/conda-forge/linux-64/proj-9.1.1-h8ffa02c_2.conda#c264aea0e16bba26afa0a0940e954492
+https://conda.anaconda.org/conda-forge/linux-64/pulseaudio-16.1-ha8d29e2_1.conda#dbfc2a8d63a43a11acf4c704e1ef9d0c
+https://conda.anaconda.org/conda-forge/noarch/pygments-2.14.0-pyhd8ed1ab_0.conda#c78cd16b11cd6a295484bd6c8f24bea1
+https://conda.anaconda.org/conda-forge/noarch/pytest-7.2.2-pyhd8ed1ab_0.conda#60958b19354e0ec295b43f6ab5cfab86
https://conda.anaconda.org/conda-forge/noarch/python-dateutil-2.8.2-pyhd8ed1ab_0.tar.bz2#dd999d1cc9f79e67dbb855c8924c7984
https://conda.anaconda.org/conda-forge/linux-64/python-stratify-0.2.post0-py38h26c90d9_3.tar.bz2#6e7902b0e96f42fa1b73daa5f65dd669
-https://conda.anaconda.org/conda-forge/linux-64/pywavelets-1.3.0-py38h26c90d9_2.tar.bz2#d30399a3c636c75cfd3460c92effa960
-https://conda.anaconda.org/conda-forge/linux-64/scipy-1.9.3-py38h8ce737c_2.tar.bz2#dfd81898f0c6e9ee0c22305da6aa443e
-https://conda.anaconda.org/conda-forge/linux-64/shapely-1.8.5-py38hafd38ec_2.tar.bz2#8df75c6a8c1deac4e99583ec624ff327
-https://conda.anaconda.org/conda-forge/linux-64/sip-6.7.5-py38hfa26641_0.conda#7be81814bae276dc7b4c707cf1e8186b
+https://conda.anaconda.org/conda-forge/linux-64/pywavelets-1.4.1-py38h7e4f40d_0.conda#17f682c947f9cabd348e7276f00c6d85
+https://conda.anaconda.org/conda-forge/linux-64/shapely-2.0.1-py38hd07e089_0.conda#84c9262ab4057ed9f80888fcfc4bf60a
+https://conda.anaconda.org/conda-forge/linux-64/sip-6.7.7-py38h8dc9893_0.conda#ea242937718f3dacf253355e1d634535
https://conda.anaconda.org/conda-forge/noarch/typing-extensions-4.4.0-hd8ed1ab_0.tar.bz2#be969210b61b897775a0de63cd9e9026
-https://conda.anaconda.org/conda-forge/linux-64/virtualenv-20.17.0-py38h578d9bd_0.conda#d89831246b5ea571858611690c3c75a4
+https://conda.anaconda.org/conda-forge/noarch/accessible-pygments-0.0.3-pyhd8ed1ab_0.conda#9838acb5f38ac58240741f5cea70a952
https://conda.anaconda.org/conda-forge/linux-64/brotlipy-0.7.0-py38h0a891b7_1005.tar.bz2#e99e08812dfff30fdd17b3f8838e2759
https://conda.anaconda.org/conda-forge/linux-64/cf-units-3.1.1-py38h26c90d9_2.tar.bz2#0ea017e84efe45badce6c32f274dbf8e
-https://conda.anaconda.org/conda-forge/linux-64/cryptography-38.0.4-py38h80a4ca7_0.conda#d3c4698fd7475640f4d9eff8d792deac
-https://conda.anaconda.org/conda-forge/noarch/dask-core-2022.12.0-pyhd8ed1ab_0.conda#3a0f020d07998e1ae711df071f97fc19
-https://conda.anaconda.org/conda-forge/linux-64/gstreamer-1.21.2-hd4edc92_0.conda#3ae425efddb9da5fb35edda331e4dff7
-https://conda.anaconda.org/conda-forge/linux-64/harfbuzz-5.3.0-h418a68e_0.tar.bz2#888056bd4b12e110b10d4d1f29161c5e
-https://conda.anaconda.org/conda-forge/noarch/imagehash-4.3.1-pyhd8ed1ab_0.tar.bz2#132ad832787a2156be1f1b309835001a
-https://conda.anaconda.org/conda-forge/linux-64/libnetcdf-4.8.1-mpi_mpich_hcd871d9_6.tar.bz2#6cdc429ed22edb566ac4308f3da6916d
-https://conda.anaconda.org/conda-forge/linux-64/matplotlib-base-3.6.2-py38hb021067_0.tar.bz2#72422499195d8aded0dfd461c6e3e86f
-https://conda.anaconda.org/conda-forge/linux-64/pandas-1.5.2-py38h8f669ce_0.conda#dbc17622f9d159be987bd21959d5494e
-https://conda.anaconda.org/conda-forge/linux-64/pyproj-3.4.0-py38hce0a2d1_2.tar.bz2#be61a535f279bffdf7f449a654eaa19d
-https://conda.anaconda.org/conda-forge/linux-64/pyqt5-sip-12.11.0-py38hfa26641_2.tar.bz2#ad6437509a14f1e8e5b8a354f93f340c
-https://conda.anaconda.org/conda-forge/noarch/pytest-xdist-3.1.0-pyhd8ed1ab_0.conda#e82f8fb903d7c4a59c77954759c341f9
-https://conda.anaconda.org/conda-forge/noarch/setuptools-scm-7.0.5-pyhd8ed1ab_1.tar.bz2#07037fe2931871ed69b2b3d2acd5fdc6
+https://conda.anaconda.org/conda-forge/linux-64/cryptography-39.0.2-py38h3d167d9_0.conda#6c60377f8bfa325a2cd80d603627a613
+https://conda.anaconda.org/conda-forge/noarch/dask-core-2023.3.0-pyhd8ed1ab_0.conda#34437340f37faafad7a6287d3b624f60
+https://conda.anaconda.org/conda-forge/linux-64/gstreamer-1.22.0-h25f0c4b_0.conda#d764367398de61c0d5531dd912e6cc96
+https://conda.anaconda.org/conda-forge/linux-64/harfbuzz-6.0.0-h8e241bc_0.conda#448fe40d2fed88ccf4d9ded37cbb2b38
+https://conda.anaconda.org/conda-forge/noarch/importlib-resources-5.12.0-pyhd8ed1ab_0.conda#3544c818f0720c89eb16ae6940ab440b
+https://conda.anaconda.org/conda-forge/linux-64/libnetcdf-4.9.1-mpi_mpich_hf10a581_1.conda#1d235cbeed74dc63e22e41779838bec1
+https://conda.anaconda.org/conda-forge/linux-64/pandas-1.5.3-py38hdc8b05c_0.conda#5073966d63a54434d2a2fc41d325b072
+https://conda.anaconda.org/conda-forge/noarch/platformdirs-3.1.0-pyhd8ed1ab_0.conda#0b8fbdfd52918bc2f1b76feccd95c919
+https://conda.anaconda.org/conda-forge/linux-64/pyproj-3.4.1-py38h58d5fe2_1.conda#5286eaec7e93586e4ae05e7d658cd3e2
+https://conda.anaconda.org/conda-forge/linux-64/pyqt5-sip-12.11.0-py38h8dc9893_3.conda#7bb0328b4a0f857aeb432426b9a5f908
+https://conda.anaconda.org/conda-forge/noarch/pytest-xdist-3.2.0-pyhd8ed1ab_0.conda#70ab87b96126f35d1e68de2ad9fb6423
+https://conda.anaconda.org/conda-forge/noarch/setuptools-scm-7.1.0-pyhd8ed1ab_0.conda#6613dbb3b25cc648a107f33ca9f80fc1
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-napoleon-0.7-py_0.tar.bz2#0bc25ff6f2e34af63ded59692df5f749
https://conda.anaconda.org/conda-forge/linux-64/ukkonen-1.0.1-py38h43d8883_3.tar.bz2#82b3797d08a43a101b645becbb938e65
-https://conda.anaconda.org/conda-forge/linux-64/cartopy-0.21.0-py38hf6c3373_3.tar.bz2#1dc477fef9b0b1080af3e7c7ecb4aff7
-https://conda.anaconda.org/conda-forge/linux-64/gst-plugins-base-1.21.2-h3e40eee_0.conda#52cbed7e92713cf01b76445530396695
-https://conda.anaconda.org/conda-forge/noarch/identify-2.5.9-pyhd8ed1ab_0.conda#e7ecbbb61a37daed2a13de43d35d5282
-https://conda.anaconda.org/conda-forge/noarch/nc-time-axis-1.4.1-pyhd8ed1ab_0.tar.bz2#281b58948bf60a2582de9e548bcc5369
-https://conda.anaconda.org/conda-forge/linux-64/netcdf-fortran-4.6.0-mpi_mpich_hd09bd1e_1.tar.bz2#0b69750bb937cab0db14f6bcef6fd787
-https://conda.anaconda.org/conda-forge/linux-64/netcdf4-1.6.0-nompi_py38h6b4b75c_103.conda#ea3d2204fc3a7db7d831daa437a58717
-https://conda.anaconda.org/conda-forge/linux-64/pango-1.50.12-h382ae3d_0.conda#627bea5af786dbd8013ef26127d8115a
-https://conda.anaconda.org/conda-forge/noarch/pyopenssl-22.1.0-pyhd8ed1ab_0.tar.bz2#fbfa0a180d48c800f922a10a114a8632
-https://conda.anaconda.org/conda-forge/linux-64/esmf-8.2.0-mpi_mpich_h5a1934d_102.tar.bz2#bb8bdfa5e3e9e3f6ec861f05cd2ad441
+https://conda.anaconda.org/conda-forge/linux-64/gst-plugins-base-1.22.0-h4243ec0_0.conda#81c20b15d2281a1ea48eac5b4eee8cfa
+https://conda.anaconda.org/conda-forge/noarch/identify-2.5.18-pyhd8ed1ab_0.conda#e07a5691c27e65d8d3d9278c578c7771
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-base-3.7.0-py38hd6c3c57_0.conda#dd63f6486ba95c036b6bfe0b5c53d875
+https://conda.anaconda.org/conda-forge/linux-64/netcdf-fortran-4.6.0-mpi_mpich_ha3603da_3.conda#ea38e2d3c472876ff4bf6551c17a9a1a
+https://conda.anaconda.org/conda-forge/linux-64/netcdf4-1.6.2-nompi_py38h08fe49f_101.conda#d2f666fab1d9a1948928756ca8ac1824
+https://conda.anaconda.org/conda-forge/linux-64/pango-1.50.14-hd33c08f_0.conda#a8b9e35dd7be2c945b0de4fe19a7c3a9
+https://conda.anaconda.org/conda-forge/linux-64/parallelio-2.5.10-mpi_mpich_h773ea27_101.conda#0793f7cf646b9bf66d83d93394474083
+https://conda.anaconda.org/conda-forge/noarch/pyopenssl-23.0.0-pyhd8ed1ab_0.conda#d41957700e83bbb925928764cb7f8878
+https://conda.anaconda.org/conda-forge/noarch/virtualenv-20.20.0-pyhd8ed1ab_0.conda#a4c92707c28aafc95208c747db80fd3f
+https://conda.anaconda.org/conda-forge/linux-64/esmf-8.4.0-mpi_mpich_h7b33e6e_105.conda#14f813a98a4158556c50084bf8e46a78
https://conda.anaconda.org/conda-forge/linux-64/gtk2-2.24.33-h90689f9_2.tar.bz2#957a0255ab58aaf394a91725d73ab422
https://conda.anaconda.org/conda-forge/linux-64/librsvg-2.54.4-h7abd40a_0.tar.bz2#921e53675ed5ea352f022b79abab076a
-https://conda.anaconda.org/conda-forge/linux-64/pre-commit-2.20.0-py38h578d9bd_1.tar.bz2#38d9029214399e4bfc378b62b0171bf0
-https://conda.anaconda.org/conda-forge/linux-64/qt-main-5.15.6-he99da89_3.conda#b7b364a82ad3ce9e56f0bad77efa9ab1
-https://conda.anaconda.org/conda-forge/noarch/urllib3-1.26.13-pyhd8ed1ab_0.conda#3078ef2359efd6ecadbc7e085c5e0592
-https://conda.anaconda.org/conda-forge/linux-64/esmpy-8.2.0-mpi_mpich_py38h9147699_101.tar.bz2#5a9de1dec507b6614150a77d1aabf257
-https://conda.anaconda.org/conda-forge/linux-64/graphviz-6.0.2-h99bc08f_0.conda#8f247587d1520a2bbc6f79a821b74c07
-https://conda.anaconda.org/conda-forge/linux-64/pyqt-5.15.7-py38h7492b6b_2.tar.bz2#cfa725eff634872f90dcd5ebf8e8dc1a
-https://conda.anaconda.org/conda-forge/noarch/requests-2.28.1-pyhd8ed1ab_1.tar.bz2#089382ee0e2dc2eae33a04cc3c2bddb0
-https://conda.anaconda.org/conda-forge/linux-64/matplotlib-3.6.2-py38h578d9bd_0.tar.bz2#e1a19f0d4686a701d4a4acce2b625acb
+https://conda.anaconda.org/conda-forge/noarch/nc-time-axis-1.4.1-pyhd8ed1ab_0.tar.bz2#281b58948bf60a2582de9e548bcc5369
+https://conda.anaconda.org/conda-forge/linux-64/pre-commit-3.1.1-py38h578d9bd_0.conda#df03025924528b963abdf907258b6852
+https://conda.anaconda.org/conda-forge/linux-64/qt-main-5.15.8-h5d23da1_6.conda#59c73debd9405771690ddbbad6c57b69
+https://conda.anaconda.org/conda-forge/noarch/urllib3-1.26.14-pyhd8ed1ab_0.conda#01f33ad2e0aaf6b5ba4add50dad5ad29
+https://conda.anaconda.org/conda-forge/linux-64/esmpy-8.4.0-mpi_mpich_py38h4407c66_102.conda#9a5c841acef11d7e4f0bf98cbc6308b3
+https://conda.anaconda.org/conda-forge/linux-64/graphviz-7.1.0-h2e5815a_0.conda#e7ecda996c443142a0e9c379f3b28e48
+https://conda.anaconda.org/conda-forge/linux-64/pyqt-5.15.7-py38ha0d8c90_3.conda#e965dc172d67920d058ac2b3a0e27565
+https://conda.anaconda.org/conda-forge/noarch/requests-2.28.2-pyhd8ed1ab_0.conda#11d178fc55199482ee48d6812ea83983
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-3.7.0-py38h578d9bd_0.conda#7fb6ab52eb5de5023445561d86dbd602
+https://conda.anaconda.org/conda-forge/noarch/pooch-1.7.0-pyhd8ed1ab_0.conda#eb2e0fc33ad0d04b51f9280360c13c1e
https://conda.anaconda.org/conda-forge/noarch/sphinx-4.5.0-pyh6c4a22f_0.tar.bz2#46b38d88c4270ff9ba78a89c83c66345
-https://conda.anaconda.org/conda-forge/noarch/pydata-sphinx-theme-0.8.1-pyhd8ed1ab_0.tar.bz2#7d8390ec71225ea9841b276552fdffba
+https://conda.anaconda.org/conda-forge/noarch/pydata-sphinx-theme-0.13.0-pyhd8ed1ab_0.conda#d9916a8dd3f0ee9c795109ee76c5dee6
+https://conda.anaconda.org/conda-forge/linux-64/scipy-1.10.1-py38h10c12cc_0.conda#1cbc47bb9a600ce4a49d8da797d375bf
https://conda.anaconda.org/conda-forge/noarch/sphinx-copybutton-0.5.0-pyhd8ed1ab_0.tar.bz2#4c969cdd5191306c269490f7ff236d9c
https://conda.anaconda.org/conda-forge/noarch/sphinx-gallery-0.11.1-pyhd8ed1ab_0.tar.bz2#729254314a5d178eefca50acbc2687b8
https://conda.anaconda.org/conda-forge/noarch/sphinx-panels-0.6.0-pyhd8ed1ab_0.tar.bz2#6eec6480601f5d15babf9c3b3987f34a
+https://conda.anaconda.org/conda-forge/linux-64/cartopy-0.21.1-py38h3d2c718_0.conda#55ba6e3a49c4293302262286a49607d8
+https://conda.anaconda.org/conda-forge/noarch/imagehash-4.3.1-pyhd8ed1ab_0.tar.bz2#132ad832787a2156be1f1b309835001a
diff --git a/requirements/ci/nox.lock/py39-linux-64.lock b/requirements/ci/nox.lock/py39-linux-64.lock
index f3f071da34..4d7ce0a576 100644
--- a/requirements/ci/nox.lock/py39-linux-64.lock
+++ b/requirements/ci/nox.lock/py39-linux-64.lock
@@ -1,6 +1,6 @@
# Generated by conda-lock.
# platform: linux-64
-# input_hash: 8720b47771aff1b233330a6562a535e5ad3a153a023d02d4dc71b383a25796a3
+# input_hash: 23dff964b0b7254aa6b68bd471a7276f62e9eaa86280f550ef4f34a2022201e0
@EXPLICIT
https://conda.anaconda.org/conda-forge/linux-64/_libgcc_mutex-0.1-conda_forge.tar.bz2#d7c89558ba9fa0495403155b64376d81
https://conda.anaconda.org/conda-forge/linux-64/ca-certificates-2022.12.7-ha878542_0.conda#ff9f73d45c4a07d6f424495288a26080
@@ -8,7 +8,7 @@ https://conda.anaconda.org/conda-forge/noarch/font-ttf-dejavu-sans-mono-2.37-hab
https://conda.anaconda.org/conda-forge/noarch/font-ttf-inconsolata-3.000-h77eed37_0.tar.bz2#34893075a5c9e55cdafac56607368fc6
https://conda.anaconda.org/conda-forge/noarch/font-ttf-source-code-pro-2.038-h77eed37_0.tar.bz2#4d59c254e01d9cde7957100457e2d5fb
https://conda.anaconda.org/conda-forge/noarch/font-ttf-ubuntu-0.83-hab24e00_0.tar.bz2#19410c3df09dfb12d1206132a1d357c5
-https://conda.anaconda.org/conda-forge/linux-64/ld_impl_linux-64-2.39-hcc3a1bd_1.conda#737be0d34c22d24432049ab7a3214de4
+https://conda.anaconda.org/conda-forge/linux-64/ld_impl_linux-64-2.40-h41732ed_0.conda#7aca3059a1729aa76c597603f10b0dd3
https://conda.anaconda.org/conda-forge/linux-64/libgfortran5-12.2.0-h337968e_19.tar.bz2#164b4b1acaedc47ee7e658ae6b308ca3
https://conda.anaconda.org/conda-forge/linux-64/libstdcxx-ng-12.2.0-h46fd767_19.tar.bz2#1030b1f38c129f2634eae026f704fe60
https://conda.anaconda.org/conda-forge/linux-64/mpi-1.0-mpich.tar.bz2#c1fcff3417b5a22bbc4cf6e8c23648cf
@@ -33,13 +33,14 @@ https://conda.anaconda.org/conda-forge/linux-64/giflib-5.2.1-h36c2ea0_2.tar.bz2#
https://conda.anaconda.org/conda-forge/linux-64/graphite2-1.3.13-h58526e2_1001.tar.bz2#8c54672728e8ec6aa6db90cf2806d220
https://conda.anaconda.org/conda-forge/linux-64/gstreamer-orc-0.4.33-h166bdaf_0.tar.bz2#879c93426c9d0b84a9de4513fbce5f4f
https://conda.anaconda.org/conda-forge/linux-64/icu-70.1-h27087fc_0.tar.bz2#87473a15119779e021c314249d4b4aed
-https://conda.anaconda.org/conda-forge/linux-64/jpeg-9e-h166bdaf_2.tar.bz2#ee8b844357a0946870901c7c6f418268
+https://conda.anaconda.org/conda-forge/linux-64/jpeg-9e-h0b41bf4_3.conda#c7a069243e1fbe9a556ed2ec030e6407
https://conda.anaconda.org/conda-forge/linux-64/keyutils-1.6.1-h166bdaf_0.tar.bz2#30186d27e2c9fa62b45fb1476b7200e3
https://conda.anaconda.org/conda-forge/linux-64/lame-3.100-h166bdaf_1003.tar.bz2#a8832b479f93521a9e7b5b743803be51
https://conda.anaconda.org/conda-forge/linux-64/lerc-4.0.0-h27087fc_0.tar.bz2#76bbff344f0134279f225174e9064c8f
+https://conda.anaconda.org/conda-forge/linux-64/libaec-1.0.6-hcb278e6_1.conda#0f683578378cddb223e7fd24f785ab2a
https://conda.anaconda.org/conda-forge/linux-64/libbrotlicommon-1.0.9-h166bdaf_8.tar.bz2#9194c9bf9428035a05352d031462eae4
https://conda.anaconda.org/conda-forge/linux-64/libdb-6.2.32-h9c3ff4c_0.tar.bz2#3f3258d8f841fbac63b36b75bdac1afd
-https://conda.anaconda.org/conda-forge/linux-64/libdeflate-1.14-h166bdaf_0.tar.bz2#fc84a0446e4e4fb882e78d786cfb9734
+https://conda.anaconda.org/conda-forge/linux-64/libdeflate-1.17-h0b41bf4_0.conda#5cc781fd91968b11a8a7fdbee0982676
https://conda.anaconda.org/conda-forge/linux-64/libev-4.33-h516909a_1.tar.bz2#6f8720dff19e17ce5d48cfe7f3d2f0a3
https://conda.anaconda.org/conda-forge/linux-64/libffi-3.4.2-h7f98852_5.tar.bz2#d645c6d2ac96843a2bfaccd2d62b3ac3
https://conda.anaconda.org/conda-forge/linux-64/libiconv-1.17-h166bdaf_0.tar.bz2#b62b52da46c39ee2bc3c162ac7f1804d
@@ -48,17 +49,17 @@ https://conda.anaconda.org/conda-forge/linux-64/libnsl-2.0.0-h7f98852_0.tar.bz2#
https://conda.anaconda.org/conda-forge/linux-64/libogg-1.3.4-h7f98852_1.tar.bz2#6e8cc2173440d77708196c5b93771680
https://conda.anaconda.org/conda-forge/linux-64/libopenblas-0.3.21-pthreads_h78a6416_3.tar.bz2#8c5963a49b6035c40646a763293fbb35
https://conda.anaconda.org/conda-forge/linux-64/libopus-1.3.1-h7f98852_1.tar.bz2#15345e56d527b330e1cacbdf58676e8f
-https://conda.anaconda.org/conda-forge/linux-64/libtool-2.4.6-h9c3ff4c_1008.tar.bz2#16e143a1ed4b4fd169536373957f6fee
-https://conda.anaconda.org/conda-forge/linux-64/libudev1-252-h166bdaf_0.tar.bz2#174243089ec111479298a5b7099b64b5
+https://conda.anaconda.org/conda-forge/linux-64/libtool-2.4.7-h27087fc_0.conda#f204c8ba400ec475452737094fb81d52
+https://conda.anaconda.org/conda-forge/linux-64/libudev1-253-h0b41bf4_0.conda#6c2addbd9aa4ee47c76d50c9f0df8cd6
https://conda.anaconda.org/conda-forge/linux-64/libuuid-2.32.1-h7f98852_1000.tar.bz2#772d69f030955d9646d3d0eaf21d859d
https://conda.anaconda.org/conda-forge/linux-64/libwebp-base-1.2.4-h166bdaf_0.tar.bz2#ac2ccf7323d21f2994e4d1f5da664f37
https://conda.anaconda.org/conda-forge/linux-64/libzlib-1.2.13-h166bdaf_4.tar.bz2#f3f9de449d32ca9b9c66a22863c96f41
-https://conda.anaconda.org/conda-forge/linux-64/lz4-c-1.9.3-h9c3ff4c_1.tar.bz2#fbe97e8fa6f275d7c76a09e795adc3e6
-https://conda.anaconda.org/conda-forge/linux-64/mpg123-1.31.1-h27087fc_0.tar.bz2#0af513b75f78a701a152568a31303bdf
+https://conda.anaconda.org/conda-forge/linux-64/lz4-c-1.9.4-hcb278e6_0.conda#318b08df404f9c9be5712aaa5a6f0bb0
+https://conda.anaconda.org/conda-forge/linux-64/mpg123-1.31.2-hcb278e6_0.conda#08efb1e1813f1a151b7a945b972a049b
https://conda.anaconda.org/conda-forge/linux-64/mpich-4.0.3-h846660c_100.tar.bz2#50d66bb751cfa71ee2a48b2d3eb90ac1
https://conda.anaconda.org/conda-forge/linux-64/ncurses-6.3-h27087fc_1.tar.bz2#4acfc691e64342b9dae57cf2adc63238
https://conda.anaconda.org/conda-forge/linux-64/nspr-4.35-h27087fc_0.conda#da0ec11a6454ae19bff5b02ed881a2b1
-https://conda.anaconda.org/conda-forge/linux-64/openssl-3.0.7-h0b41bf4_1.conda#7adaac6ff98219bcb99b45e408b80f4e
+https://conda.anaconda.org/conda-forge/linux-64/openssl-3.0.8-h0b41bf4_0.conda#e043403cd18faf815bf7705ab6c1e092
https://conda.anaconda.org/conda-forge/linux-64/pixman-0.40.0-h36c2ea0_0.tar.bz2#660e72c82f2e75a6b3fe6a6e75c79f19
https://conda.anaconda.org/conda-forge/linux-64/pthread-stubs-0.4-h36c2ea0_1001.tar.bz2#22dad4df6e8630e8dff2428f6f6a7036
https://conda.anaconda.org/conda-forge/linux-64/xorg-kbproto-1.0.7-h7f98852_1002.tar.bz2#4b230e8381279d76131116660f5a241a
@@ -66,12 +67,12 @@ https://conda.anaconda.org/conda-forge/linux-64/xorg-libice-1.0.10-h7f98852_0.ta
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxau-1.0.9-h7f98852_0.tar.bz2#bf6f803a544f26ebbdc3bfff272eb179
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxdmcp-1.1.3-h7f98852_0.tar.bz2#be93aabceefa2fac576e971aef407908
https://conda.anaconda.org/conda-forge/linux-64/xorg-renderproto-0.11.1-h7f98852_1002.tar.bz2#06feff3d2634e3097ce2fe681474b534
-https://conda.anaconda.org/conda-forge/linux-64/xorg-xextproto-7.3.0-h7f98852_1002.tar.bz2#1e15f6ad85a7d743a2ac68dae6c82b98
+https://conda.anaconda.org/conda-forge/linux-64/xorg-xextproto-7.3.0-h0b41bf4_1003.conda#bce9f945da8ad2ae9b1d7165a64d0f87
https://conda.anaconda.org/conda-forge/linux-64/xorg-xproto-7.0.31-h7f98852_1007.tar.bz2#b4a4381d54784606820704f7b5f05a15
-https://conda.anaconda.org/conda-forge/linux-64/xxhash-0.8.0-h7f98852_3.tar.bz2#52402c791f35e414e704b7a113f99605
+https://conda.anaconda.org/conda-forge/linux-64/xxhash-0.8.1-h0b41bf4_0.conda#e9c3bcf0e0c719431abec8ca447eee27
https://conda.anaconda.org/conda-forge/linux-64/xz-5.2.6-h166bdaf_0.tar.bz2#2161070d867d1b1204ea749c8eec4ef0
https://conda.anaconda.org/conda-forge/linux-64/yaml-0.2.5-h7f98852_2.tar.bz2#4cb3ad778ec2d5a7acbdf254eb1c42ae
-https://conda.anaconda.org/conda-forge/linux-64/jack-1.9.21-h583fa2b_2.conda#7b36a10b58964d4444fcba44244710c5
+https://conda.anaconda.org/conda-forge/linux-64/jack-1.9.22-h11f4161_0.conda#504fa9e712b99494a9cf4630e3ca7d78
https://conda.anaconda.org/conda-forge/linux-64/libblas-3.9.0-16_linux64_openblas.tar.bz2#d9b7a8639171f6c6fa0a983edabcfe2b
https://conda.anaconda.org/conda-forge/linux-64/libbrotlidec-1.0.9-h166bdaf_8.tar.bz2#4ae4d7795d33e02bd20f6b23d91caf82
https://conda.anaconda.org/conda-forge/linux-64/libbrotlienc-1.0.9-h166bdaf_8.tar.bz2#04bac51ba35ea023dc48af73c1c88c25
@@ -79,8 +80,8 @@ https://conda.anaconda.org/conda-forge/linux-64/libcap-2.66-ha37c62d_0.tar.bz2#2
https://conda.anaconda.org/conda-forge/linux-64/libedit-3.1.20191231-he28a2e2_2.tar.bz2#4d331e44109e3f0e19b4cb8f9b82f3e1
https://conda.anaconda.org/conda-forge/linux-64/libevent-2.1.10-h28343ad_4.tar.bz2#4a049fc560e00e43151dc51368915fdd
https://conda.anaconda.org/conda-forge/linux-64/libflac-1.4.2-h27087fc_0.tar.bz2#7daf72d8e2a8e848e11d63ed6d1026e0
-https://conda.anaconda.org/conda-forge/linux-64/libgpg-error-1.45-hc0c96e0_0.tar.bz2#839aeb24ab885a7b902247a6d943d02f
-https://conda.anaconda.org/conda-forge/linux-64/libnghttp2-1.47.0-hff17c54_1.tar.bz2#2b7dbfa6988a41f9d23ba6d4f0e1d74e
+https://conda.anaconda.org/conda-forge/linux-64/libgpg-error-1.46-h620e276_0.conda#27e745f6f2e4b757e95dd7225fbe6bdb
+https://conda.anaconda.org/conda-forge/linux-64/libnghttp2-1.52.0-h61bc06f_0.conda#613955a50485812985c059e7b269f42e
https://conda.anaconda.org/conda-forge/linux-64/libpng-1.6.39-h753d276_0.conda#e1c890aebdebbfbf87e2c917187b4416
https://conda.anaconda.org/conda-forge/linux-64/libsqlite-3.40.0-h753d276_0.tar.bz2#2e5f9a37d487e1019fd4d8113adb2f9f
https://conda.anaconda.org/conda-forge/linux-64/libssh2-1.10.0-hf14f497_3.tar.bz2#d85acad4b47dff4e3def14a769a97906
@@ -88,78 +89,77 @@ https://conda.anaconda.org/conda-forge/linux-64/libvorbis-1.3.7-h9c3ff4c_0.tar.b
https://conda.anaconda.org/conda-forge/linux-64/libxcb-1.13-h7f98852_1004.tar.bz2#b3653fdc58d03face9724f602218a904
https://conda.anaconda.org/conda-forge/linux-64/libxml2-2.10.3-h7463322_0.tar.bz2#3b933ea47ef8f330c4c068af25fcd6a8
https://conda.anaconda.org/conda-forge/linux-64/libzip-1.9.2-hc929e4a_1.tar.bz2#5b122b50e738c4be5c3f2899f010d7cf
-https://conda.anaconda.org/conda-forge/linux-64/mysql-common-8.0.31-h26416b9_0.tar.bz2#6c531bc30d49ae75b9c7c7f65bd62e3c
+https://conda.anaconda.org/conda-forge/linux-64/mysql-common-8.0.32-ha901b37_0.conda#6a39818710235826181e104aada40c75
https://conda.anaconda.org/conda-forge/linux-64/pcre2-10.40-hc3806b6_0.tar.bz2#69e2c796349cd9b273890bee0febfe1b
https://conda.anaconda.org/conda-forge/linux-64/readline-8.1.2-h0f457ee_0.tar.bz2#db2ebbe2943aae81ed051a6a9af8e0fa
https://conda.anaconda.org/conda-forge/linux-64/tk-8.6.12-h27826a3_0.tar.bz2#5b8c42eb62e9fc961af70bdd6a26e168
https://conda.anaconda.org/conda-forge/linux-64/udunits2-2.2.28-hc3e0081_0.tar.bz2#d4c341e0379c31e9e781d4f204726867
https://conda.anaconda.org/conda-forge/linux-64/xorg-libsm-1.2.3-hd9c2040_1000.tar.bz2#9e856f78d5c80d5a78f61e72d1d473a3
https://conda.anaconda.org/conda-forge/linux-64/zlib-1.2.13-h166bdaf_4.tar.bz2#4b11e365c0275b808be78b30f904e295
-https://conda.anaconda.org/conda-forge/linux-64/zstd-1.5.2-h6239696_4.tar.bz2#adcf0be7897e73e312bd24353b613f74
+https://conda.anaconda.org/conda-forge/linux-64/zstd-1.5.2-h3eb15da_6.conda#6b63daed8feeca47be78f323e793d555
https://conda.anaconda.org/conda-forge/linux-64/brotli-bin-1.0.9-h166bdaf_8.tar.bz2#e5613f2bc717e9945840ff474419b8e4
https://conda.anaconda.org/conda-forge/linux-64/freetype-2.12.1-hca18f0e_1.conda#e1232042de76d24539a436d37597eb06
https://conda.anaconda.org/conda-forge/linux-64/hdf4-4.2.15-h9772cbc_5.tar.bz2#ee08782aff2ff9b3291c967fa6bc7336
-https://conda.anaconda.org/conda-forge/linux-64/krb5-1.19.3-h08a2579_0.tar.bz2#d25e05e7ee0e302b52d24491db4891eb
+https://conda.anaconda.org/conda-forge/linux-64/krb5-1.20.1-h81ceb04_0.conda#89a41adce7106749573d883b2f657d78
https://conda.anaconda.org/conda-forge/linux-64/libcblas-3.9.0-16_linux64_openblas.tar.bz2#20bae26d0a1db73f758fc3754cab4719
https://conda.anaconda.org/conda-forge/linux-64/libgcrypt-1.10.1-h166bdaf_0.tar.bz2#f967fc95089cd247ceed56eda31de3a9
https://conda.anaconda.org/conda-forge/linux-64/libglib-2.74.1-h606061b_1.tar.bz2#ed5349aa96776e00b34eccecf4a948fe
https://conda.anaconda.org/conda-forge/linux-64/liblapack-3.9.0-16_linux64_openblas.tar.bz2#955d993f41f9354bf753d29864ea20ad
-https://conda.anaconda.org/conda-forge/linux-64/libllvm15-15.0.6-h63197d8_0.conda#201168ef66095bbd565e124ee2c56a20
-https://conda.anaconda.org/conda-forge/linux-64/libsndfile-1.1.0-hcb278e6_1.conda#d7a07b1f5974bce4735112aaef0c1467
-https://conda.anaconda.org/conda-forge/linux-64/libtiff-4.4.0-h55922b4_4.tar.bz2#901791f0ec7cddc8714e76e273013a91
-https://conda.anaconda.org/conda-forge/linux-64/libxkbcommon-1.0.3-he3ba5ed_0.tar.bz2#f9dbabc7e01c459ed7a1d1d64b206e9b
-https://conda.anaconda.org/conda-forge/linux-64/mysql-libs-8.0.31-hbc51c84_0.tar.bz2#da9633eee814d4e910fe42643a356315
-https://conda.anaconda.org/conda-forge/linux-64/nss-3.82-he02c5a1_0.conda#f8d7f11d19e4cb2207eab159fd4c0152
-https://conda.anaconda.org/conda-forge/linux-64/python-3.9.15-hba424b6_0_cpython.conda#7b9485fce17fac2dd4aca6117a9936c2
+https://conda.anaconda.org/conda-forge/linux-64/libllvm15-15.0.7-hadd5161_0.conda#70cbb0c2033665f2a7339bf0ec51a67f
+https://conda.anaconda.org/conda-forge/linux-64/libsndfile-1.2.0-hb75c966_0.conda#c648d19cd9c8625898d5d370414de7c7
+https://conda.anaconda.org/conda-forge/linux-64/libtiff-4.5.0-h6adf6a1_2.conda#2e648a34072eb39d7c4fc2a9981c5f0c
+https://conda.anaconda.org/conda-forge/linux-64/libxkbcommon-1.5.0-h79f4944_0.conda#3f67368c9b0e77a693acad193310baf1
+https://conda.anaconda.org/conda-forge/linux-64/mysql-libs-8.0.32-hd7da12d_0.conda#b05d7ea8b76f1172d5fe4f30e03277ea
+https://conda.anaconda.org/conda-forge/linux-64/nss-3.88-he45b914_0.conda#d7a81dfb99ad8fbb88872fb7ec646e6c
+https://conda.anaconda.org/conda-forge/linux-64/python-3.9.16-h2782a2a_0_cpython.conda#95c9b7c96a7fd7342e0c9d0a917b8f78
https://conda.anaconda.org/conda-forge/linux-64/sqlite-3.40.0-h4ff8645_0.tar.bz2#bb11803129cbbb53ed56f9506ff74145
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-0.4.0-h166bdaf_0.tar.bz2#384e7fcb3cd162ba3e4aed4b687df566
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-keysyms-0.4.0-h166bdaf_0.tar.bz2#637054603bb7594302e3bf83f0a99879
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-renderutil-0.3.9-h166bdaf_0.tar.bz2#732e22f1741bccea861f5668cf7342a7
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-wm-0.4.1-h166bdaf_0.tar.bz2#0a8e20a8aef954390b9481a527421a8c
-https://conda.anaconda.org/conda-forge/linux-64/xorg-libx11-1.7.2-h7f98852_0.tar.bz2#12a61e640b8894504326aadafccbb790
-https://conda.anaconda.org/conda-forge/noarch/alabaster-0.7.12-py_0.tar.bz2#2489a97287f90176ecdc3ca982b4b0a0
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libx11-1.8.4-h0b41bf4_0.conda#ea8fbfeb976ac49cbeb594e985393514
+https://conda.anaconda.org/conda-forge/noarch/alabaster-0.7.13-pyhd8ed1ab_0.conda#06006184e203b61d3525f90de394471e
https://conda.anaconda.org/conda-forge/linux-64/antlr-python-runtime-4.7.2-py39hf3d152e_1003.tar.bz2#5e8330e806e50bd6137ebd125f4bc1bb
https://conda.anaconda.org/conda-forge/linux-64/atk-1.0-2.38.0-hd4edc92_1.tar.bz2#6c72ec3e660a51736913ef6ea68c454b
-https://conda.anaconda.org/conda-forge/noarch/attrs-22.1.0-pyh71513ae_1.tar.bz2#6d3ccbc56256204925bfa8378722792f
+https://conda.anaconda.org/conda-forge/noarch/attrs-22.2.0-pyh71513ae_0.conda#8b76db7818a4e401ed4486c4c1635cd9
https://conda.anaconda.org/conda-forge/linux-64/brotli-1.0.9-h166bdaf_8.tar.bz2#2ff08978892a3e8b954397c461f18418
https://conda.anaconda.org/conda-forge/noarch/certifi-2022.12.7-pyhd8ed1ab_0.conda#fb9addc3db06e56abe03e0e9f21a63e6
https://conda.anaconda.org/conda-forge/noarch/cfgv-3.3.1-pyhd8ed1ab_0.tar.bz2#ebb5f5f7dc4f1a3780ef7ea7738db08c
https://conda.anaconda.org/conda-forge/noarch/charset-normalizer-2.1.1-pyhd8ed1ab_0.tar.bz2#c1d5b294fbf9a795dec349a6f4d8be8e
https://conda.anaconda.org/conda-forge/noarch/click-8.1.3-unix_pyhd8ed1ab_2.tar.bz2#20e4087407c7cb04a40817114b333dbf
-https://conda.anaconda.org/conda-forge/noarch/cloudpickle-2.2.0-pyhd8ed1ab_0.tar.bz2#a6cf47b09786423200d7982d1faa19eb
+https://conda.anaconda.org/conda-forge/noarch/cloudpickle-2.2.1-pyhd8ed1ab_0.conda#b325bfc4cff7d7f8a868f1f7ecc4ed16
https://conda.anaconda.org/conda-forge/noarch/colorama-0.4.6-pyhd8ed1ab_0.tar.bz2#3faab06a954c2a04039983f2c4a50d99
https://conda.anaconda.org/conda-forge/noarch/cycler-0.11.0-pyhd8ed1ab_0.tar.bz2#a50559fad0affdbb33729a68669ca1cb
https://conda.anaconda.org/conda-forge/linux-64/dbus-1.13.6-h5008d03_3.tar.bz2#ecfff944ba3960ecb334b9a2663d708d
https://conda.anaconda.org/conda-forge/noarch/distlib-0.3.6-pyhd8ed1ab_0.tar.bz2#b65b4d50dbd2d50fa0aeac367ec9eed7
https://conda.anaconda.org/conda-forge/linux-64/docutils-0.17.1-py39hf3d152e_3.tar.bz2#3caf51fb6a259d377f05d6913193b11c
-https://conda.anaconda.org/conda-forge/noarch/exceptiongroup-1.0.4-pyhd8ed1ab_0.tar.bz2#e0734d1f12de77f9daca98bda3428733
+https://conda.anaconda.org/conda-forge/noarch/exceptiongroup-1.1.0-pyhd8ed1ab_0.conda#a385c3e8968b4cf8fbc426ace915fd1a
https://conda.anaconda.org/conda-forge/noarch/execnet-1.9.0-pyhd8ed1ab_0.tar.bz2#0e521f7a5e60d508b121d38b04874fb2
-https://conda.anaconda.org/conda-forge/noarch/filelock-3.8.2-pyhd8ed1ab_0.conda#0f09c2bc17ddd8732be8e5b99297c7ce
-https://conda.anaconda.org/conda-forge/linux-64/fontconfig-2.14.1-hc2a2eb6_0.tar.bz2#78415f0180a8d9c5bcc47889e00d5fb1
-https://conda.anaconda.org/conda-forge/noarch/fsspec-2022.11.0-pyhd8ed1ab_0.tar.bz2#eb919f2119a6db5d0192f9e9c3711572
-https://conda.anaconda.org/conda-forge/linux-64/gdk-pixbuf-2.42.8-hff1cb4f_1.tar.bz2#a61c6312192e7c9de71548a6706a21e6
+https://conda.anaconda.org/conda-forge/noarch/filelock-3.9.0-pyhd8ed1ab_0.conda#1addc115923d646ca19ed90edc413506
+https://conda.anaconda.org/conda-forge/linux-64/fontconfig-2.14.2-h14ed4e7_0.conda#0f69b688f52ff6da70bccb7ff7001d1d
+https://conda.anaconda.org/conda-forge/noarch/fsspec-2023.1.0-pyhd8ed1ab_0.conda#44f6828b8f7cc3433d68d1d1c0e9add2
+https://conda.anaconda.org/conda-forge/linux-64/gdk-pixbuf-2.42.10-h05c8ddd_0.conda#1a109126a43003d65b39c1cad656bc9b
https://conda.anaconda.org/conda-forge/linux-64/glib-tools-2.74.1-h6239696_1.tar.bz2#5f442e6bc9d89ba236eb25a25c5c2815
https://conda.anaconda.org/conda-forge/linux-64/gts-0.7.6-h64030ff_2.tar.bz2#112eb9b5b93f0c02e59aea4fd1967363
https://conda.anaconda.org/conda-forge/noarch/idna-3.4-pyhd8ed1ab_0.tar.bz2#34272b248891bddccc64479f9a7fffed
https://conda.anaconda.org/conda-forge/noarch/imagesize-1.4.1-pyhd8ed1ab_0.tar.bz2#7de5386c8fea29e76b303f37dde4c352
-https://conda.anaconda.org/conda-forge/noarch/iniconfig-1.1.1-pyh9f0ad1d_0.tar.bz2#39161f81cc5e5ca45b8226fbb06c6905
+https://conda.anaconda.org/conda-forge/noarch/iniconfig-2.0.0-pyhd8ed1ab_0.conda#f800d2da156d08e289b14e87e43c1ae5
https://conda.anaconda.org/conda-forge/noarch/iris-sample-data-2.4.0-pyhd8ed1ab_0.tar.bz2#18ee9c07cf945a33f92caf1ee3d23ad9
https://conda.anaconda.org/conda-forge/linux-64/kiwisolver-1.4.4-py39hf939315_1.tar.bz2#41679a052a8ce841c74df1ebc802e411
-https://conda.anaconda.org/conda-forge/linux-64/lcms2-2.14-h6ed2654_0.tar.bz2#dcc588839de1445d90995a0a2c4f3a39
-https://conda.anaconda.org/conda-forge/linux-64/libclang13-15.0.6-default_h3a83d3e_0.conda#535dd0ca1dcb165b6a8ffa10d01945fe
-https://conda.anaconda.org/conda-forge/linux-64/libcups-2.3.3-h3e49a29_2.tar.bz2#3b88f1d0fe2580594d58d7e44d664617
-https://conda.anaconda.org/conda-forge/linux-64/libcurl-7.86.0-h2283fc2_1.tar.bz2#fdca8cd67ec2676f90a70ac73a32538b
-https://conda.anaconda.org/conda-forge/linux-64/libpq-15.1-h67c24c5_1.conda#e1389a8d9a907133b3e6483c2807d243
+https://conda.anaconda.org/conda-forge/linux-64/lcms2-2.14-hfd0df8a_1.conda#c2566c2ea5f153ddd6bf4acaf7547d97
+https://conda.anaconda.org/conda-forge/linux-64/libclang13-15.0.7-default_h3e3d535_1.conda#a3a0f7a6f0885f5e1e0ec691566afb77
+https://conda.anaconda.org/conda-forge/linux-64/libcups-2.3.3-h36d4200_3.conda#c9f4416a34bc91e0eb029f912c68f81f
+https://conda.anaconda.org/conda-forge/linux-64/libcurl-7.88.1-hdc1c0ab_0.conda#81eaeb3b35163c8e90e57532bc93754d
+https://conda.anaconda.org/conda-forge/linux-64/libpq-15.2-hb675445_0.conda#4654b17eccaba55b8581d6b9c77f53cc
https://conda.anaconda.org/conda-forge/linux-64/libsystemd0-252-h2a991cd_0.tar.bz2#3c5ae9f61f663b3d5e1bf7f7da0c85f5
-https://conda.anaconda.org/conda-forge/linux-64/libwebp-1.2.4-h522a892_0.tar.bz2#802e43f480122a85ae6a34c1909f8f98
+https://conda.anaconda.org/conda-forge/linux-64/libwebp-1.2.4-h1daa5a0_1.conda#77003f63d1763c1e6569a02c1742c9f4
https://conda.anaconda.org/conda-forge/noarch/locket-1.0.0-pyhd8ed1ab_0.tar.bz2#91e27ef3d05cc772ce627e51cff111c4
-https://conda.anaconda.org/conda-forge/linux-64/markupsafe-2.1.1-py39hb9d737c_2.tar.bz2#c678e07e7862b3157fb9f6d908233ffa
+https://conda.anaconda.org/conda-forge/linux-64/markupsafe-2.1.2-py39h72bdee0_0.conda#35514f5320206df9f4661c138c02e1c1
https://conda.anaconda.org/conda-forge/linux-64/mpi4py-3.1.4-py39h32b9844_0.tar.bz2#b035b507f55bb6a967d86d4b7e059437
https://conda.anaconda.org/conda-forge/noarch/munkres-1.1.4-pyh9f0ad1d_0.tar.bz2#2ba8498c1018c1e9c61eb99b973dfe19
-https://conda.anaconda.org/conda-forge/linux-64/numpy-1.23.5-py39h3d75532_0.conda#ea5d332e361eb72c2593cf79559bc0ec
-https://conda.anaconda.org/conda-forge/linux-64/openjpeg-2.5.0-h7d73246_1.tar.bz2#a11b4df9271a8d7917686725aa04c8f2
-https://conda.anaconda.org/conda-forge/noarch/packaging-22.0-pyhd8ed1ab_0.conda#0e8e1bd93998978fc3125522266d12db
-https://conda.anaconda.org/conda-forge/noarch/platformdirs-2.6.0-pyhd8ed1ab_0.conda#b1b2ab02d1ece1719f7fa002ad4bc70d
+https://conda.anaconda.org/conda-forge/linux-64/numpy-1.24.2-py39h7360e5f_0.conda#757070dc7cc33003254888808cd34f1e
+https://conda.anaconda.org/conda-forge/linux-64/openjpeg-2.5.0-hfec8fc6_2.conda#5ce6a42505c6e9e6151c54c3ec8d68ea
+https://conda.anaconda.org/conda-forge/noarch/packaging-23.0-pyhd8ed1ab_0.conda#1ff2e3ca41f0ce16afec7190db28288b
https://conda.anaconda.org/conda-forge/noarch/pluggy-1.0.0-pyhd8ed1ab_5.tar.bz2#7d301a0d25f424d96175f810935f0da9
https://conda.anaconda.org/conda-forge/noarch/ply-3.11-py_1.tar.bz2#7205635cd71531943440fbfe3b6b5727
https://conda.anaconda.org/conda-forge/linux-64/psutil-5.9.4-py39hb9d737c_0.tar.bz2#12184951da572828fb986b06ffb63eed
@@ -167,16 +167,16 @@ https://conda.anaconda.org/conda-forge/noarch/pycparser-2.21-pyhd8ed1ab_0.tar.bz
https://conda.anaconda.org/conda-forge/noarch/pyparsing-3.0.9-pyhd8ed1ab_0.tar.bz2#e8fbc1b54b25f4b08281467bc13b70cc
https://conda.anaconda.org/conda-forge/noarch/pyshp-2.3.1-pyhd8ed1ab_0.tar.bz2#92a889dc236a5197612bc85bee6d7174
https://conda.anaconda.org/conda-forge/noarch/pysocks-1.7.1-pyha2e5f31_6.tar.bz2#2a7de29fb590ca14b5243c4c812c8025
-https://conda.anaconda.org/conda-forge/linux-64/python-xxhash-3.0.0-py39hb9d737c_2.tar.bz2#b643f1e19306b75a6013d77228156076
-https://conda.anaconda.org/conda-forge/noarch/pytz-2022.6-pyhd8ed1ab_0.tar.bz2#b1f26ad83328e486910ef7f6e81dc061
+https://conda.anaconda.org/conda-forge/linux-64/python-xxhash-3.2.0-py39h72bdee0_0.conda#18927f971926b7271600368de71de557
+https://conda.anaconda.org/conda-forge/noarch/pytz-2022.7.1-pyhd8ed1ab_0.conda#f59d49a7b464901cf714b9e7984d01a2
https://conda.anaconda.org/conda-forge/linux-64/pyyaml-6.0-py39hb9d737c_5.tar.bz2#ef9db3c38ae7275f6b14491cfe61a248
-https://conda.anaconda.org/conda-forge/noarch/setuptools-65.5.1-pyhd8ed1ab_0.tar.bz2#cfb8dc4d9d285ca5fb1177b9dd450e33
+https://conda.anaconda.org/conda-forge/noarch/setuptools-67.4.0-pyhd8ed1ab_0.conda#c6f4b87020c72e2700e3e94c1fc93b70
https://conda.anaconda.org/conda-forge/noarch/six-1.16.0-pyh6c4a22f_0.tar.bz2#e5f25f8dbc060e9a8d912e432202afc2
https://conda.anaconda.org/conda-forge/noarch/snowballstemmer-2.2.0-pyhd8ed1ab_0.tar.bz2#4d22a9315e78c6827f806065957d566e
https://conda.anaconda.org/conda-forge/noarch/soupsieve-2.3.2.post1-pyhd8ed1ab_0.tar.bz2#146f4541d643d48fc8a75cacf69f03ae
-https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-applehelp-1.0.2-py_0.tar.bz2#20b2eaeaeea4ef9a9a0d99770620fd09
+https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-applehelp-1.0.4-pyhd8ed1ab_0.conda#5a31a7d564f551d0e6dff52fd8cb5b16
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-devhelp-1.0.2-py_0.tar.bz2#68e01cac9d38d0e717cd5c87bc3d2cc9
-https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-htmlhelp-2.0.0-pyhd8ed1ab_0.tar.bz2#77dad82eb9c8c1525ff7953e0756d708
+https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-htmlhelp-2.0.1-pyhd8ed1ab_0.conda#6c8c4d6eb2325e59290ac6dbbeacd5f0
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-jsmath-1.0.1-py_0.tar.bz2#67cd9d9c0382d37479b4d306c369a2d4
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-qthelp-1.0.3-py_0.tar.bz2#d01180388e6d1838c3e1ad029590aa7a
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-serializinghtml-1.1.5-pyhd8ed1ab_2.tar.bz2#9ff55a0901cf952f05c654394de76bf7
@@ -188,78 +188,84 @@ https://conda.anaconda.org/conda-forge/noarch/typing_extensions-4.4.0-pyha770c72
https://conda.anaconda.org/conda-forge/linux-64/unicodedata2-15.0.0-py39hb9d737c_0.tar.bz2#230d65004135bf312504a1bbcb0c7a08
https://conda.anaconda.org/conda-forge/noarch/wheel-0.38.4-pyhd8ed1ab_0.tar.bz2#c829cfb8cb826acb9de0ac1a2df0a940
https://conda.anaconda.org/conda-forge/linux-64/xcb-util-image-0.4.0-h166bdaf_0.tar.bz2#c9b568bd804cb2903c6be6f5f68182e4
-https://conda.anaconda.org/conda-forge/linux-64/xorg-libxext-1.3.4-h7f98852_1.tar.bz2#536cc5db4d0a3ba0630541aec064b5e4
+https://conda.anaconda.org/conda-forge/linux-64/xorg-libxext-1.3.4-h0b41bf4_2.conda#82b6df12252e6f32402b96dacc656fec
https://conda.anaconda.org/conda-forge/linux-64/xorg-libxrender-0.9.10-h7f98852_1003.tar.bz2#f59c1242cc1dd93e72c2ee2b360979eb
-https://conda.anaconda.org/conda-forge/noarch/zipp-3.11.0-pyhd8ed1ab_0.conda#09b5b885341697137879a4f039a9e5a1
-https://conda.anaconda.org/conda-forge/noarch/babel-2.11.0-pyhd8ed1ab_0.tar.bz2#2ea70fde8d581ba9425a761609eed6ba
-https://conda.anaconda.org/conda-forge/noarch/beautifulsoup4-4.11.1-pyha770c72_0.tar.bz2#eeec8814bd97b2681f708bb127478d7d
+https://conda.anaconda.org/conda-forge/noarch/zipp-3.15.0-pyhd8ed1ab_0.conda#13018819ca8f5b7cc675a8faf1f5fedf
+https://conda.anaconda.org/conda-forge/noarch/babel-2.12.1-pyhd8ed1ab_1.conda#ac432e732804a81ddcf29c92ead57cde
+https://conda.anaconda.org/conda-forge/noarch/beautifulsoup4-4.11.2-pyha770c72_0.conda#88b59f6989f0ed5ab3433af0b82555e1
https://conda.anaconda.org/conda-forge/linux-64/cairo-1.16.0-ha61ee94_1014.tar.bz2#d1a88f3ed5b52e1024b80d4bcd26a7a0
-https://conda.anaconda.org/conda-forge/linux-64/cffi-1.15.1-py39he91dace_2.tar.bz2#fc70a133e8162f51e363cff3b6dc741c
+https://conda.anaconda.org/conda-forge/linux-64/cffi-1.15.1-py39he91dace_3.conda#20080319ef73fbad74dcd6d62f2a3ffe
https://conda.anaconda.org/conda-forge/linux-64/cftime-1.6.2-py39h2ae25f5_1.tar.bz2#c943fb9a2818ecc5be1e0ecc8b7738f1
-https://conda.anaconda.org/conda-forge/linux-64/contourpy-1.0.6-py39hf939315_0.tar.bz2#fb3f77fe25042c20c51974fcfe72f797
-https://conda.anaconda.org/conda-forge/linux-64/curl-7.86.0-h2283fc2_1.tar.bz2#9d4149760567cb232691cce2d8ccc21f
+https://conda.anaconda.org/conda-forge/linux-64/contourpy-1.0.7-py39h4b4f3f3_0.conda#c5387f3fb1f5b8b71e1c865fc55f4951
+https://conda.anaconda.org/conda-forge/linux-64/curl-7.88.1-hdc1c0ab_0.conda#1968e4fef727858ac04746560e820928
https://conda.anaconda.org/conda-forge/linux-64/fonttools-4.38.0-py39hb9d737c_1.tar.bz2#3f2d104f2fefdd5e8a205dd3aacbf1d7
https://conda.anaconda.org/conda-forge/linux-64/glib-2.74.1-h6239696_1.tar.bz2#f3220a9e9d3abcbfca43419a219df7e4
-https://conda.anaconda.org/conda-forge/linux-64/hdf5-1.12.2-mpi_mpich_h5d83325_0.tar.bz2#6b5c2d276f306df759cfbdb0f41c4db9
-https://conda.anaconda.org/conda-forge/noarch/importlib-metadata-5.1.0-pyha770c72_0.conda#46a62e35b9ae515cf0e49afc7fe0e7ef
+https://conda.anaconda.org/conda-forge/linux-64/hdf5-1.12.2-mpi_mpich_h5d83325_1.conda#811c4d55cf17b42336ffa314239717b0
+https://conda.anaconda.org/conda-forge/noarch/importlib-metadata-6.0.0-pyha770c72_0.conda#691644becbcdca9f73243450b1c63e62
+https://conda.anaconda.org/conda-forge/noarch/importlib_resources-5.12.0-pyhd8ed1ab_0.conda#e5fd2260a231ee63b6969f4801082f2b
https://conda.anaconda.org/conda-forge/noarch/jinja2-3.1.2-pyhd8ed1ab_1.tar.bz2#c8490ed5c70966d232fdd389d0dbed37
-https://conda.anaconda.org/conda-forge/linux-64/libclang-15.0.6-default_h2e3cab8_0.conda#1b2cee49acc5b03c73ad0f68bfe04bb8
-https://conda.anaconda.org/conda-forge/linux-64/libgd-2.3.3-h18fbbfe_3.tar.bz2#ea9758cf553476ddf75c789fdd239dc5
+https://conda.anaconda.org/conda-forge/linux-64/libclang-15.0.7-default_had23c3d_1.conda#36c65ed73b7c92589bd9562ef8a6023d
+https://conda.anaconda.org/conda-forge/linux-64/libgd-2.3.3-h5aea950_4.conda#82ef57611ace65b59db35a9687264572
https://conda.anaconda.org/conda-forge/linux-64/mo_pack-0.2.0-py39h2ae25f5_1008.tar.bz2#d90acb3804f16c63eb6726652e4e25b3
https://conda.anaconda.org/conda-forge/noarch/nodeenv-1.7.0-pyhd8ed1ab_0.tar.bz2#fbe1182f650c04513046d6894046cd6c
https://conda.anaconda.org/conda-forge/noarch/partd-1.3.0-pyhd8ed1ab_0.tar.bz2#af8c82d121e63082926062d61d9abb54
-https://conda.anaconda.org/conda-forge/linux-64/pillow-9.2.0-py39hf3a2cdf_3.tar.bz2#2bd111c38da69056e5fe25a51b832eba
-https://conda.anaconda.org/conda-forge/noarch/pip-22.3.1-pyhd8ed1ab_0.tar.bz2#da66f2851b9836d3a7c5190082a45f7d
+https://conda.anaconda.org/conda-forge/linux-64/pillow-9.4.0-py39h2320bf1_1.conda#d2f79132b9c8e416058a4cd84ef27b3d
+https://conda.anaconda.org/conda-forge/noarch/pip-23.0.1-pyhd8ed1ab_0.conda#8025ca83b8ba5430b640b83917c2a6f7
https://conda.anaconda.org/conda-forge/noarch/pockets-0.9.1-py_0.tar.bz2#1b52f0c42e8077e5a33e00fe72269364
-https://conda.anaconda.org/conda-forge/linux-64/proj-9.1.0-h93bde94_0.tar.bz2#255c7204dda39747c3ba380d28b026d7
-https://conda.anaconda.org/conda-forge/linux-64/pulseaudio-16.1-h126f2b6_0.tar.bz2#e4b74b33e13dd146e7d8b5078fc9ad30
-https://conda.anaconda.org/conda-forge/noarch/pygments-2.13.0-pyhd8ed1ab_0.tar.bz2#9f478e8eedd301008b5f395bad0caaed
-https://conda.anaconda.org/conda-forge/noarch/pytest-7.2.0-pyhd8ed1ab_2.tar.bz2#ac82c7aebc282e6ac0450fca012ca78c
+https://conda.anaconda.org/conda-forge/linux-64/proj-9.1.1-h8ffa02c_2.conda#c264aea0e16bba26afa0a0940e954492
+https://conda.anaconda.org/conda-forge/linux-64/pulseaudio-16.1-ha8d29e2_1.conda#dbfc2a8d63a43a11acf4c704e1ef9d0c
+https://conda.anaconda.org/conda-forge/noarch/pygments-2.14.0-pyhd8ed1ab_0.conda#c78cd16b11cd6a295484bd6c8f24bea1
+https://conda.anaconda.org/conda-forge/noarch/pytest-7.2.2-pyhd8ed1ab_0.conda#60958b19354e0ec295b43f6ab5cfab86
https://conda.anaconda.org/conda-forge/noarch/python-dateutil-2.8.2-pyhd8ed1ab_0.tar.bz2#dd999d1cc9f79e67dbb855c8924c7984
https://conda.anaconda.org/conda-forge/linux-64/python-stratify-0.2.post0-py39h2ae25f5_3.tar.bz2#bcc7de3bb458a198b598ac1f75bf37e3
-https://conda.anaconda.org/conda-forge/linux-64/pywavelets-1.3.0-py39h2ae25f5_2.tar.bz2#234ad9828eca1caf0f2fdcb4a24ad816
-https://conda.anaconda.org/conda-forge/linux-64/scipy-1.9.3-py39hddc5342_2.tar.bz2#0615ac8191c6ccf7d40860aff645f774
-https://conda.anaconda.org/conda-forge/linux-64/shapely-1.8.5-py39h76a96b7_2.tar.bz2#10bea68a9dd064b703743d210e679408
-https://conda.anaconda.org/conda-forge/linux-64/sip-6.7.5-py39h5a03fae_0.conda#c3eb463691a8b93f1c381a9e56ecad9a
+https://conda.anaconda.org/conda-forge/linux-64/pywavelets-1.4.1-py39h389d5f1_0.conda#9eeb2b2549f836ca196c6cbd22344122
+https://conda.anaconda.org/conda-forge/linux-64/shapely-2.0.1-py39hc9151fd_0.conda#d26cc40830285883abaa766a7f7798bf
+https://conda.anaconda.org/conda-forge/linux-64/sip-6.7.7-py39h227be39_0.conda#7d9a35091552af3655151f164ddd64a3
https://conda.anaconda.org/conda-forge/noarch/typing-extensions-4.4.0-hd8ed1ab_0.tar.bz2#be969210b61b897775a0de63cd9e9026
-https://conda.anaconda.org/conda-forge/linux-64/virtualenv-20.17.0-py39hf3d152e_0.conda#a6f9ae6d84b4b233968e20a707935462
+https://conda.anaconda.org/conda-forge/noarch/accessible-pygments-0.0.3-pyhd8ed1ab_0.conda#9838acb5f38ac58240741f5cea70a952
https://conda.anaconda.org/conda-forge/linux-64/brotlipy-0.7.0-py39hb9d737c_1005.tar.bz2#a639fdd9428d8b25f8326a3838d54045
https://conda.anaconda.org/conda-forge/linux-64/cf-units-3.1.1-py39h2ae25f5_2.tar.bz2#b3b4aab96d1c4ed394d6f4b9146699d4
-https://conda.anaconda.org/conda-forge/linux-64/cryptography-38.0.4-py39h3ccb8fc_0.conda#dee37fde01f9bbc53ec421199d7b17cf
-https://conda.anaconda.org/conda-forge/noarch/dask-core-2022.12.0-pyhd8ed1ab_0.conda#3a0f020d07998e1ae711df071f97fc19
-https://conda.anaconda.org/conda-forge/linux-64/gstreamer-1.21.2-hd4edc92_0.conda#3ae425efddb9da5fb35edda331e4dff7
-https://conda.anaconda.org/conda-forge/linux-64/harfbuzz-5.3.0-h418a68e_0.tar.bz2#888056bd4b12e110b10d4d1f29161c5e
-https://conda.anaconda.org/conda-forge/noarch/imagehash-4.3.1-pyhd8ed1ab_0.tar.bz2#132ad832787a2156be1f1b309835001a
-https://conda.anaconda.org/conda-forge/linux-64/libnetcdf-4.8.1-mpi_mpich_hcd871d9_6.tar.bz2#6cdc429ed22edb566ac4308f3da6916d
-https://conda.anaconda.org/conda-forge/linux-64/matplotlib-base-3.6.2-py39hf9fd14e_0.tar.bz2#78ce32061e0be12deb8e0f11ffb76906
-https://conda.anaconda.org/conda-forge/linux-64/pandas-1.5.2-py39h4661b88_0.conda#e17e50269c268d79478956a262a9fe13
-https://conda.anaconda.org/conda-forge/linux-64/pyproj-3.4.0-py39h14a8356_2.tar.bz2#5d93c781338ff274a0b3dc3d901e19a6
-https://conda.anaconda.org/conda-forge/linux-64/pyqt5-sip-12.11.0-py39h5a03fae_2.tar.bz2#306f1a018668f06a0bd89350a3f62c07
-https://conda.anaconda.org/conda-forge/noarch/pytest-xdist-3.1.0-pyhd8ed1ab_0.conda#e82f8fb903d7c4a59c77954759c341f9
-https://conda.anaconda.org/conda-forge/noarch/setuptools-scm-7.0.5-pyhd8ed1ab_1.tar.bz2#07037fe2931871ed69b2b3d2acd5fdc6
+https://conda.anaconda.org/conda-forge/linux-64/cryptography-39.0.2-py39h079d5ae_0.conda#c492b565817a019f025c7d17b57ef479
+https://conda.anaconda.org/conda-forge/noarch/dask-core-2023.3.0-pyhd8ed1ab_0.conda#34437340f37faafad7a6287d3b624f60
+https://conda.anaconda.org/conda-forge/linux-64/gstreamer-1.22.0-h25f0c4b_0.conda#d764367398de61c0d5531dd912e6cc96
+https://conda.anaconda.org/conda-forge/linux-64/harfbuzz-6.0.0-h8e241bc_0.conda#448fe40d2fed88ccf4d9ded37cbb2b38
+https://conda.anaconda.org/conda-forge/noarch/importlib-resources-5.12.0-pyhd8ed1ab_0.conda#3544c818f0720c89eb16ae6940ab440b
+https://conda.anaconda.org/conda-forge/linux-64/libnetcdf-4.9.1-mpi_mpich_hf10a581_1.conda#1d235cbeed74dc63e22e41779838bec1
+https://conda.anaconda.org/conda-forge/linux-64/pandas-1.5.3-py39h2ad29b5_0.conda#3ea96adbbc2a66fa45178102a9cfbecc
+https://conda.anaconda.org/conda-forge/noarch/platformdirs-3.1.0-pyhd8ed1ab_0.conda#0b8fbdfd52918bc2f1b76feccd95c919
+https://conda.anaconda.org/conda-forge/linux-64/pyproj-3.4.1-py39hf14cbfd_1.conda#67766c515601b3ee1514072d6fd060bb
+https://conda.anaconda.org/conda-forge/linux-64/pyqt5-sip-12.11.0-py39h227be39_3.conda#9e381db00691e26bcf670c3586397be1
+https://conda.anaconda.org/conda-forge/noarch/pytest-xdist-3.2.0-pyhd8ed1ab_0.conda#70ab87b96126f35d1e68de2ad9fb6423
+https://conda.anaconda.org/conda-forge/noarch/setuptools-scm-7.1.0-pyhd8ed1ab_0.conda#6613dbb3b25cc648a107f33ca9f80fc1
https://conda.anaconda.org/conda-forge/noarch/sphinxcontrib-napoleon-0.7-py_0.tar.bz2#0bc25ff6f2e34af63ded59692df5f749
https://conda.anaconda.org/conda-forge/linux-64/ukkonen-1.0.1-py39hf939315_3.tar.bz2#0f11bcdf9669a5ae0f39efd8c830209a
-https://conda.anaconda.org/conda-forge/linux-64/cartopy-0.21.0-py39h91bfd65_3.tar.bz2#7d10a2e14c08f383baae00e77bf890e5
-https://conda.anaconda.org/conda-forge/linux-64/gst-plugins-base-1.21.2-h3e40eee_0.conda#52cbed7e92713cf01b76445530396695
-https://conda.anaconda.org/conda-forge/noarch/identify-2.5.9-pyhd8ed1ab_0.conda#e7ecbbb61a37daed2a13de43d35d5282
-https://conda.anaconda.org/conda-forge/noarch/nc-time-axis-1.4.1-pyhd8ed1ab_0.tar.bz2#281b58948bf60a2582de9e548bcc5369
-https://conda.anaconda.org/conda-forge/linux-64/netcdf-fortran-4.6.0-mpi_mpich_hd09bd1e_1.tar.bz2#0b69750bb937cab0db14f6bcef6fd787
-https://conda.anaconda.org/conda-forge/linux-64/netcdf4-1.6.0-nompi_py39h94a714e_103.conda#ee29e7176b5854fa09ec17b101945401
-https://conda.anaconda.org/conda-forge/linux-64/pango-1.50.12-h382ae3d_0.conda#627bea5af786dbd8013ef26127d8115a
-https://conda.anaconda.org/conda-forge/noarch/pyopenssl-22.1.0-pyhd8ed1ab_0.tar.bz2#fbfa0a180d48c800f922a10a114a8632
-https://conda.anaconda.org/conda-forge/linux-64/esmf-8.2.0-mpi_mpich_h5a1934d_102.tar.bz2#bb8bdfa5e3e9e3f6ec861f05cd2ad441
+https://conda.anaconda.org/conda-forge/linux-64/gst-plugins-base-1.22.0-h4243ec0_0.conda#81c20b15d2281a1ea48eac5b4eee8cfa
+https://conda.anaconda.org/conda-forge/noarch/identify-2.5.18-pyhd8ed1ab_0.conda#e07a5691c27e65d8d3d9278c578c7771
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-base-3.7.0-py39he190548_0.conda#62d6ddd9e534f4d325d12470cc4961ab
+https://conda.anaconda.org/conda-forge/linux-64/netcdf-fortran-4.6.0-mpi_mpich_ha3603da_3.conda#ea38e2d3c472876ff4bf6551c17a9a1a
+https://conda.anaconda.org/conda-forge/linux-64/netcdf4-1.6.2-nompi_py39h8b3a7bc_101.conda#16e186c6b8e60ffa3ed58e0c78ea1b9e
+https://conda.anaconda.org/conda-forge/linux-64/pango-1.50.14-hd33c08f_0.conda#a8b9e35dd7be2c945b0de4fe19a7c3a9
+https://conda.anaconda.org/conda-forge/linux-64/parallelio-2.5.10-mpi_mpich_h773ea27_101.conda#0793f7cf646b9bf66d83d93394474083
+https://conda.anaconda.org/conda-forge/noarch/pyopenssl-23.0.0-pyhd8ed1ab_0.conda#d41957700e83bbb925928764cb7f8878
+https://conda.anaconda.org/conda-forge/noarch/virtualenv-20.20.0-pyhd8ed1ab_0.conda#a4c92707c28aafc95208c747db80fd3f
+https://conda.anaconda.org/conda-forge/linux-64/esmf-8.4.0-mpi_mpich_h7b33e6e_105.conda#14f813a98a4158556c50084bf8e46a78
https://conda.anaconda.org/conda-forge/linux-64/gtk2-2.24.33-h90689f9_2.tar.bz2#957a0255ab58aaf394a91725d73ab422
https://conda.anaconda.org/conda-forge/linux-64/librsvg-2.54.4-h7abd40a_0.tar.bz2#921e53675ed5ea352f022b79abab076a
-https://conda.anaconda.org/conda-forge/linux-64/pre-commit-2.20.0-py39hf3d152e_1.tar.bz2#921f8a7c2a16d18d7168fdac88b2adfe
-https://conda.anaconda.org/conda-forge/linux-64/qt-main-5.15.6-he99da89_3.conda#b7b364a82ad3ce9e56f0bad77efa9ab1
-https://conda.anaconda.org/conda-forge/noarch/urllib3-1.26.13-pyhd8ed1ab_0.conda#3078ef2359efd6ecadbc7e085c5e0592
-https://conda.anaconda.org/conda-forge/linux-64/esmpy-8.2.0-mpi_mpich_py39h8bb458d_101.tar.bz2#347f324dd99dfb0b1479a466213b55bf
-https://conda.anaconda.org/conda-forge/linux-64/graphviz-6.0.2-h99bc08f_0.conda#8f247587d1520a2bbc6f79a821b74c07
-https://conda.anaconda.org/conda-forge/linux-64/pyqt-5.15.7-py39h18e9c17_2.tar.bz2#384809c51fb2adc04773f6fa097cd051
-https://conda.anaconda.org/conda-forge/noarch/requests-2.28.1-pyhd8ed1ab_1.tar.bz2#089382ee0e2dc2eae33a04cc3c2bddb0
-https://conda.anaconda.org/conda-forge/linux-64/matplotlib-3.6.2-py39hf3d152e_0.tar.bz2#03225b4745d1dee7bb19d81e41c773a0
+https://conda.anaconda.org/conda-forge/noarch/nc-time-axis-1.4.1-pyhd8ed1ab_0.tar.bz2#281b58948bf60a2582de9e548bcc5369
+https://conda.anaconda.org/conda-forge/linux-64/pre-commit-3.1.1-py39hf3d152e_0.conda#17994a38cb9daeb1beecacec5885745c
+https://conda.anaconda.org/conda-forge/linux-64/qt-main-5.15.8-h5d23da1_6.conda#59c73debd9405771690ddbbad6c57b69
+https://conda.anaconda.org/conda-forge/noarch/urllib3-1.26.14-pyhd8ed1ab_0.conda#01f33ad2e0aaf6b5ba4add50dad5ad29
+https://conda.anaconda.org/conda-forge/linux-64/esmpy-8.4.0-mpi_mpich_py39h3088dd8_102.conda#a022e48c8b12bc56083bcce841978519
+https://conda.anaconda.org/conda-forge/linux-64/graphviz-7.1.0-h2e5815a_0.conda#e7ecda996c443142a0e9c379f3b28e48
+https://conda.anaconda.org/conda-forge/linux-64/pyqt-5.15.7-py39h5c7b992_3.conda#19e30314fe824605750da905febb8ee6
+https://conda.anaconda.org/conda-forge/noarch/requests-2.28.2-pyhd8ed1ab_0.conda#11d178fc55199482ee48d6812ea83983
+https://conda.anaconda.org/conda-forge/linux-64/matplotlib-3.7.0-py39hf3d152e_0.conda#0967228e228ebeded6a36a6f4d5509ed
+https://conda.anaconda.org/conda-forge/noarch/pooch-1.7.0-pyhd8ed1ab_0.conda#eb2e0fc33ad0d04b51f9280360c13c1e
https://conda.anaconda.org/conda-forge/noarch/sphinx-4.5.0-pyh6c4a22f_0.tar.bz2#46b38d88c4270ff9ba78a89c83c66345
-https://conda.anaconda.org/conda-forge/noarch/pydata-sphinx-theme-0.8.1-pyhd8ed1ab_0.tar.bz2#7d8390ec71225ea9841b276552fdffba
+https://conda.anaconda.org/conda-forge/noarch/pydata-sphinx-theme-0.13.0-pyhd8ed1ab_0.conda#d9916a8dd3f0ee9c795109ee76c5dee6
+https://conda.anaconda.org/conda-forge/linux-64/scipy-1.10.1-py39h7360e5f_0.conda#7584d1bc5499d25eccfd24a7f656e3ee
https://conda.anaconda.org/conda-forge/noarch/sphinx-copybutton-0.5.0-pyhd8ed1ab_0.tar.bz2#4c969cdd5191306c269490f7ff236d9c
https://conda.anaconda.org/conda-forge/noarch/sphinx-gallery-0.11.1-pyhd8ed1ab_0.tar.bz2#729254314a5d178eefca50acbc2687b8
https://conda.anaconda.org/conda-forge/noarch/sphinx-panels-0.6.0-pyhd8ed1ab_0.tar.bz2#6eec6480601f5d15babf9c3b3987f34a
+https://conda.anaconda.org/conda-forge/linux-64/cartopy-0.21.1-py39h6e7ad6e_0.conda#7cb72bd5b1e7c5a23a062db90889356b
+https://conda.anaconda.org/conda-forge/noarch/imagehash-4.3.1-pyhd8ed1ab_0.tar.bz2#132ad832787a2156be1f1b309835001a
diff --git a/requirements/ci/py310.yml b/requirements/ci/py310.yml
index d79015c055..e3bada6596 100644
--- a/requirements/ci/py310.yml
+++ b/requirements/ci/py310.yml
@@ -16,7 +16,7 @@ dependencies:
- cftime >=1.5
- dask-core >=2.26
- matplotlib >=3.5
- - netcdf4 <1.6.1
+ - netcdf4
- numpy >=1.19
- python-xxhash
- pyproj
@@ -39,6 +39,7 @@ dependencies:
- pre-commit
- psutil
- pytest
+ - pytest-cov
- pytest-xdist
- requests
@@ -48,7 +49,7 @@ dependencies:
- sphinx-copybutton
- sphinx-gallery >=0.11.0
- sphinx-panels
- - pydata-sphinx-theme
+ - pydata-sphinx-theme >=0.13.0
# Temporary minimum pins.
# See https://github.com/SciTools/iris/pull/5051
diff --git a/requirements/ci/py38.yml b/requirements/ci/py38.yml
index cd04fa94d0..271598f2ea 100644
--- a/requirements/ci/py38.yml
+++ b/requirements/ci/py38.yml
@@ -16,7 +16,7 @@ dependencies:
- cftime >=1.5
- dask-core >=2.26
- matplotlib >=3.5
- - netcdf4 <1.6.1
+ - netcdf4
- numpy >=1.19
- python-xxhash
- pyproj
@@ -48,5 +48,5 @@ dependencies:
- sphinx-copybutton
- sphinx-gallery >=0.11.0
- sphinx-panels
- - pydata-sphinx-theme
+ - pydata-sphinx-theme >=0.13.0
diff --git a/requirements/ci/py39.yml b/requirements/ci/py39.yml
index 9fec76cfde..349784ec46 100644
--- a/requirements/ci/py39.yml
+++ b/requirements/ci/py39.yml
@@ -16,7 +16,7 @@ dependencies:
- cftime >=1.5
- dask-core >=2.26
- matplotlib >=3.5
- - netcdf4 <1.6.1
+ - netcdf4
- numpy >=1.19
- python-xxhash
- pyproj
@@ -48,7 +48,7 @@ dependencies:
- sphinx-copybutton
- sphinx-gallery >=0.11.0
- sphinx-panels
- - pydata-sphinx-theme
+ - pydata-sphinx-theme >=0.13.0
# Temporary minimum pins.
# See https://github.com/SciTools/iris/pull/5051
diff --git a/setup.cfg b/setup.cfg
index 75647e6623..ba9844f5d8 100644
--- a/setup.cfg
+++ b/setup.cfg
@@ -52,7 +52,7 @@ install_requires =
cftime>=1.5.0
dask[array]>=2.26
matplotlib>=3.5
- netcdf4<1.6.1
+ netcdf4
numpy>=1.19
scipy
shapely!=1.8.3
@@ -72,9 +72,9 @@ docs =
sphinx<5
sphinx-copybutton
sphinx-gallery>=0.11.0
- sphinx_rtd_theme
sphinxcontrib-napoleon
sphinx-panels
+ pydata-sphinx-theme>=0.13.0
test =
filelock
imagehash>=4.0
 +
+ +
+ +
+