-
Notifications
You must be signed in to change notification settings - Fork 0
Tutorial
Download BUNDLE and run it on the demo dataset.
You can work with the Scala source code for BUNDLE by cloning this repository and running sbt 0.13.1. Alternatively you can just download the latest build, BUNDLE 0.1.0. Unzip the build to find:
-
bundle-0.1.0.jar, a fat jar file with two main classes:-
bundle.Bundlegenerates posterior distribution trace files. -
bundle.Distillsummarizes traces.
-
-
poly/poly.conf, a demo configuration file. -
poly/poly.tsv, data for the demo. -
poly/plot.r, an R script to plot the result.
Before running, take a look at the top of the configuration file poly/poly.conf:
data poly.tsv # Input filename for binary matrix data.
base poly # Output filenames all begin with this string.
chainlen 10000 # Run MCMC for 10000 states.
burnin 2000 # Discard first 2000 states.
infofreq 10 # Report to the terminal every 10 states.
dumpfreq 100 # Sample every 100 states.
The data file poly.tsv codes whether each of 422 etyma exist in each of 35 Central Pacific languages. Each etymon is thus considered a linguistic feature. This data was excerpted from a 2006 version of POLLEX, a very larger comparative Polynesian word list. It takes the form of a 422×35 binary matrix with tab-separated values. The first row contains labels for languages; the first column contains labels for features. The rest of the cells contain either 0 or 1.
Now run:
java -cp bundle-0.1.0.jar bundle.Bundle poly/poly.conf
This will take around ten minutes to complete. Every 10 states it will write a summary of the MCMC state to poly.out and also to the terminal. For example, it will write:
--- 2300 ---
[α 4.6750] [μρ 0.4749 λρ 0.7799] [λζ 0.8721] [K 26]
116 85 59 45 35 15 14 11 6 5 5 4 4 3 2 2 2 1 1 1 1 1 1 1 1 1
[ll -11260.12] [z -914.47] [y -6305.70] [x -4039.96]
WYA 11 FIJ 53 ROT 44 TON 99 NIU 87 EUV 83 EFU 99 SAM 96 TOK 76 ECE 80
WUV 70 WFU 60 MAE 71 MFA 59 TIK 82 ANU 55 REN 98 PIL 62 KAP 75 NKO 79
TAK 83 OJA 76 SIK 79 NGR 27 PUK 97 EAS 59 MAO 99 TUA 93 PEN 81 RAR 94
MIA 7 TAH 86 HAW 88 MQA 91 MVA 59
- First line: The MCMC is at state 2300.
- Second line:
α,μρ,λρ,λζare model hyperparameters.Kis the number of clusters, which varies from sample to sample. - Third line: These numbers are the size of each cluster, starting from the largest. They sum to 422, the number of etyma in the model.
- Fourth line:
llis the log likelihood of the model.zis the log probability of the CRP table assignment.yis the log probability of the observability matrix.xis the log probability of the data, givenzandy. In the foregoing probabilities,w,θandξhave been integrated out. Note thatll=z+y+x. - Other lines: Inferred lexicographic coverage rate for each language, normalized to 99.For example, XX etyma are attested for Anuta (ANU) in the data matrix, but in this instance the model believes that XX (of the 422 in the dataset) actually exist in Anuta. Hence it reports that Anuta has a lexicographic coverage of XX/XX = 55%.
Every 100 MCMC states, BUNDLE writes out the entire model state over several trace files:
-
poly.y/0.hex,poly.y/100.hex,poly.y/200.hex, etc.: the observability matrixy. Since it is a large binary matrix, it is written in hex, with each state in its own file. -
poly.z.log: seating assignments, one state per line. Each line denotes a set partition of the 422 etyma. For example, a line that begins1 1 2 2 2 ...means that the first two etyma are clustered together and the next three are clustered together. Each number is just a label.7 7 15 15 15 ...would mean the same thing. There is no guarantee that the label will be consistent from one state to the next. Before summarizing the clustering, one must align the labels in one state with the labels in the next state, and align those labels with the labels in the following state, etc. -
poly.log: all the other parameters, one state per line, tab separated.
When bundle.Bundle completes, run:
java -cp bundle-0.1.0.jar bundle.Distill poly/poly.conf
This does the alignment described above and produces two files:
-
poly.ez.log: a K×422 matrix, where K is the number of clusters inferred. Cell n in row k is the posterior probability that etymon n belongs in cluster k. (This probability is also conditioned on the alignment thatbundle.Distillfinds.) The row sums of this matrix are the expected number of etyma in each cluster. -
poly.th.log: a K×35 matrix. Cell l in row k is the intensity of language l in cluster k. In other words, of the etyma in cluster k, this is the fraction that exists in language l.bundle.Distillplots this in ASCII characters:
12 CLUSTERS WITH 2+ HARD MEMBERS
WFRTN EESTE WWMMT ARPKN TOSNP EMTPR MTHMM
YIOOI UFAOC UFAFI NEIAK AJIGU AAUEA IAAQV
AJTNU VUMKE VUEAK UNLPO KAKRK SOANR AHWAA
110 @. : =::
86 .:.=: ==:.. . . .
59 @@@@@ @@@@@ @@@@@ @@@@@ @@@@@ @@@@@ :@@@@
48 :. . . : @=@@@ =@===
26 :==@= :=@@: :.:.@ .= .= .
16 @= ===@: ... = ::=: @ =@@=@ @@@@@
15 ===@@ @@@@@ @@@@@ @@@@= =@=@: .: .
5 . .... ..= : : . . . .: . :.
5 @@=. .:.= @=@: :===@ @@::= @@@@@ @@@@@
6 : @@ @@=:@ . @ = = = : @ .@.@@ @ .
3 : .: .: ::.: = == @ @ .
2 @= @. @...@ @@ = @:=@ : @ :@@. =@@.
The three-letter designations for the 35 languages run along the top. Along the left are the number of ‘hard’ members in each cluster. A hard member is an etymon that belongs in a cluster with probability greater than half. In the middle, each cell represents the intensity of a language in a cluster:
| ` ` | `.` | `:` | `=` | `@`
--------- | --- | --- | --- | --- | --- Intensity | 0-20% | 20-40% | 40-60% | 60-80% | 80-100%
If you run the R script poly/plot.r (after installing the Cairo and grid packages) you will get a much prettier result:
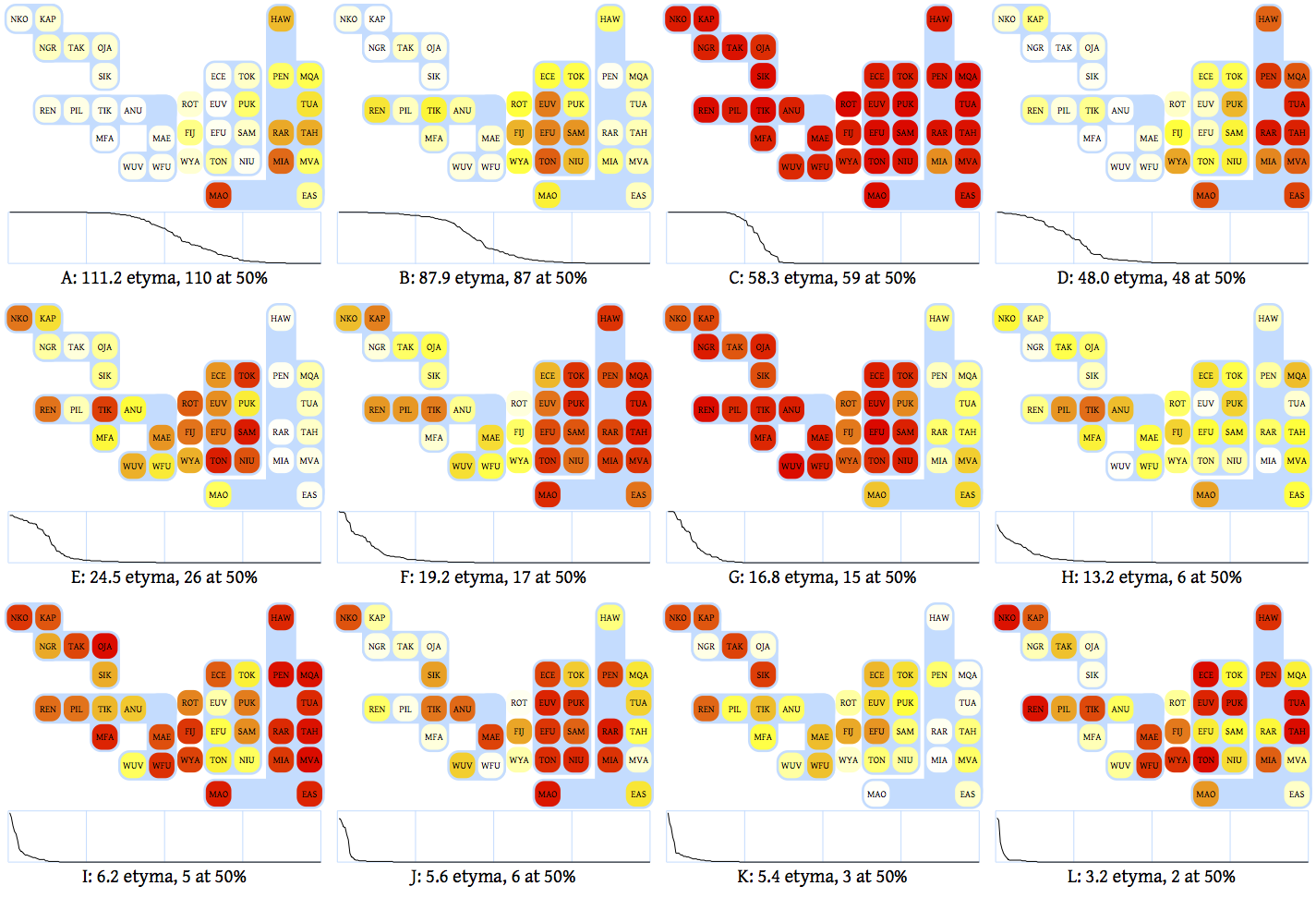
The only substantive difference is that under each cluster, the script plots the posterior probability of each etymon being in the cluster, for the 200 etyma with the highest such probabilities. This conveys the nature of membership in the cluster. Some clusters (e.g., C) have relatively sharp boundaries with respect to which etyma are in and which are out. Others (e.g., H) have quite fuzzy boundaries.
poly/plot.r is provided as an example of how to plot results. You will have to devise your own visualizations for other datasets. Feel free to modify poly/plot.r, of course.