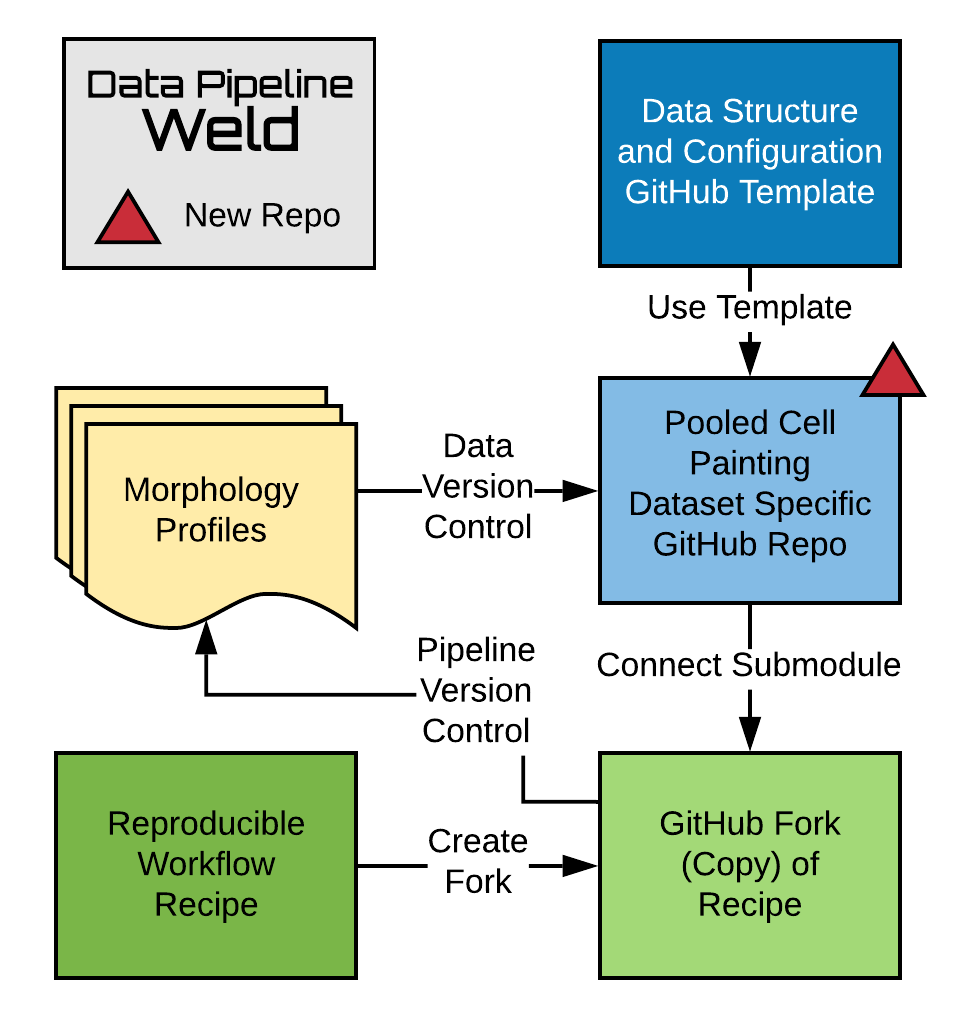
This repository was derived from a template repository located at https://github.com/broadinstitute/pooled-cell-painting-profiling-template. The purpose of the repository is to weld together a versioned data processing pipeline with versioned processed output data for a single Pooled Cell Painting experiment.
Figure 1: Data Pipeline Welding is a procedure that links together version controlled data with a version controlled processing pipeline. The procedure results in a new repository for each dataset within a Pooled Cell Painting project. The
pooled-cell-painting-profiling-reciperepository contains the data processing pipeline. Thepooled-cell-painting-profiling-template(this repo) contains recipe configuration files that must be edited for each dataset. A user's recipe fork is added to the dataset-specific repo as a Github submodule. The weld is finalized when the user prepares the recipe, outputting version controlled morphology profiles that are used for downstream biological discovery.
A note about terminology:
A batch is the data pipeline welding unit.
As the pipeline is currently written, a batch is a single plate of data and may also be referred to as a dataset.
An experiment is designed around a specific question and may contain single or multiple batches, depending on the experimental design.
A project is an encompassing term and may contain any number of experiments (and therefore any number of batches).