PyQAlloy development is a part of ULTERA Project carried under the DOE ARPA-E ULTIMATE program that aims to develop a new generation of materials for turbine blades in gas turbines and related applications. The ULTERA Project, along is led by Phases Research Lab at Penn State. As a part of it, we developed a new large-scale database of high entropy alloys (HEAs) reported in the literature along with their experimental properties. As of February 2024, the database contains around 6,500 property data points of 2,700 HEAs coming from almost 540 publications. It is currently the largest database of HEAs in the world, and while it is not publicly available we welcome collaborators who would like to use it in their research or contribute to it. ULTERA Database is not simply a dataset but features a robust set of data processing, curation, and aggregation tools we built for the last 3 years. These tools allowed us to remove around the 5-10% erroneous data we identified in datasets available in the literature, primarily with help of tools like the ones in this repository.
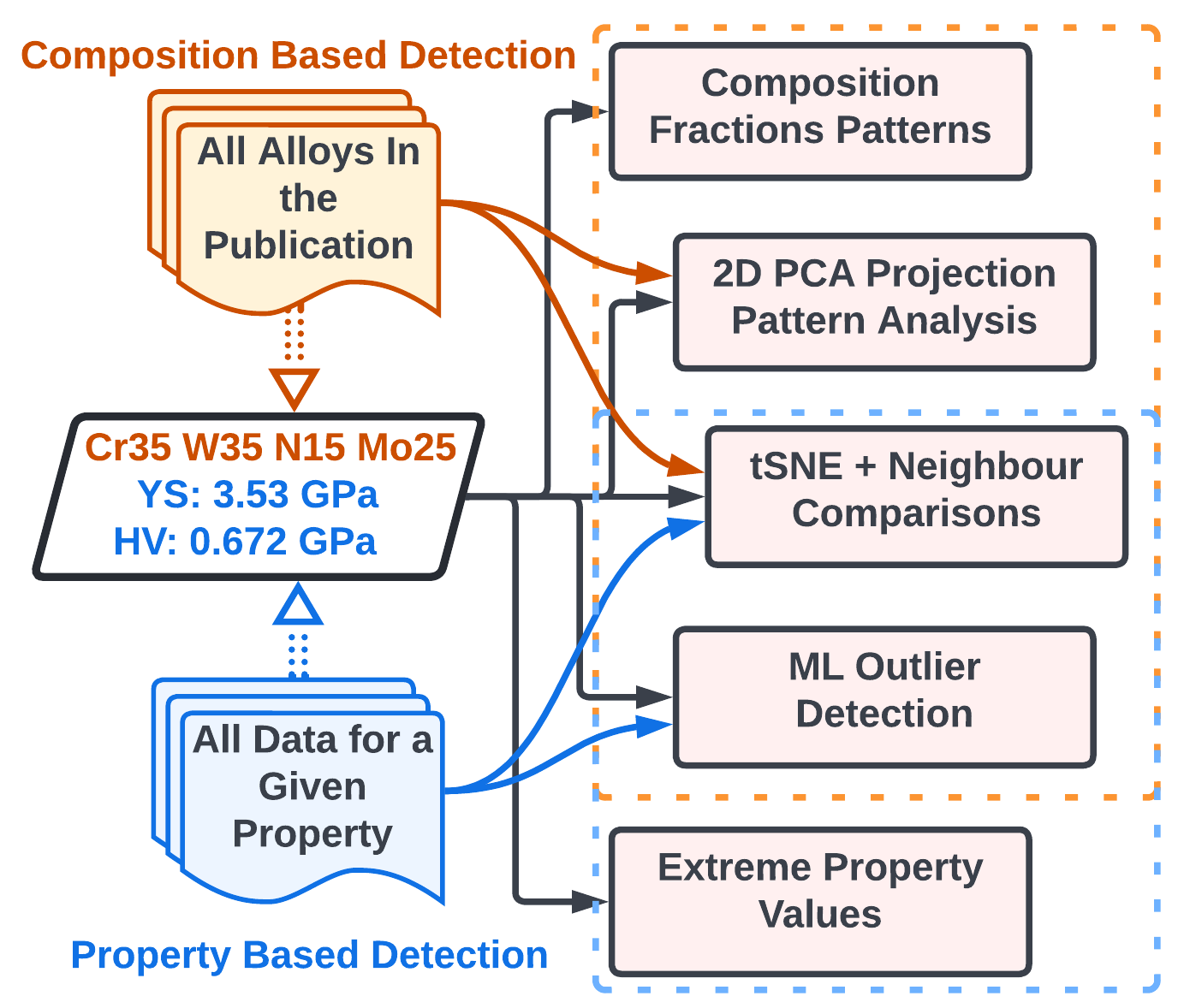
PyQAlloy is a Python package for detecting data abnormalities in datasets of arbitrary alloys, ranging from complex, concentrated solutions, i.e. High Entropy Alloys (HEAs) / Multi Principle Element Alloys (MPEAs) / Concentrated Complex Alloys (CCAs) to more traditional alloys such as steels, nickel-based superalloys, etc. As of v0.3.7, around half of the tools we developed were added here, and the rest will be published in Mid-2024. Figure below serves as a graphical abstract of our approach.
PyQAlloy is readily available on PyPI (since V0.3.5), and you can get it) with a simple:
pip install pyqalloy
Once the installation process is complete, you will be able to utilize it in your Python scripts or Jupyter notebooks.
To get a ready-to-go installation of PyQAlloy with all notebooks in this repository, it is recommended to install it in development mode - that is to clone the repository and install it in editable mode.
While not required, it is recommended to first set up a virtual environment using venv or Conda. This ensures that one of the required versions of Python (3.9+) is used and there are no dependency conflicts. If you have Conda installed on your system (see instructions at https://docs.conda.io/en/latest/miniconda.html), you can create a new environment with:
conda create -n pyqalloy python=3.9 jupyter
conda activate pyqalloy
Then, clone PyQAlloy from GitHub like
git clone https://github.com/PhasesResearchLab/PyQAlloy.git
Please note this will, by default, download
the latest development version of the software, which may not be stable. For a stable version, you can specify a version
tag after the URL with --branch <tag_name> --single-branch.
Then, move to the PyQAlloy folder and install in editable (-e) mode.
cd PyQAlloy
pip install -e .
If you are using the ULTERA Project infrastructure, now you should fill in your details into the
pyqalloy/credentials.json with name, dbKey, and dataServer fields, and you should be ready to go as the most current
stable version will be kept up-to-date with the latest stable snapshot of ULTERA! :)
You can start by going through the UserCuration.ipynb notebook. It will guide you through
all core functionalities of PyQAlloy.
You will need to set up your own MongoDB database or another tool
"pretending" to be one and fill it with data that conforms to the ULTERA schema. You can do it either manually
(instructions will be provided in the future, and we are happy to help you get started today) or by using
a snapshot of the ULTERA database subset devTools/ULTERA_sample.bson if you only want to learn how to use PyQAlloy
for now.
Start with CustomDatasetFromBSON.ipynb notebook which will show you how to create a custom MontyDB in-memory database
from a BSON file (or JSON if you prefer). Then, you can modify the UserCuration.ipynb notebook to use your custom
database and work through all exercises there.
To give a taste of PyQAlloy's interface, here is a minimal snippet that will utilize the ULTERA database and
scan it for datapoints uploaded by Adam Krajewski (who also happens to write this README) with the uncertainty of
2.1% (i.e. how much a composition can deviate from 100% to be considered a valid composition) and print the first 10
results on the fly as they are found.
from pyqalloy.curation import analysis
sC = analysis.SingleCompositionAnalyzer(name='Adam Krajewski')
sC.scanCompositionsAround100(
printOnFly=True,
resultLimit=10,
uncertainty=0.21)