This repository stores some script tools to cleanse the FAERS ASCII data. We use this dataset to build iADRs (http://iadr.csie.nuk.edu.tw/), an online web-based analytical system for detecting and analyzing suspected signals of adverse drug reactions and drug-interactions.
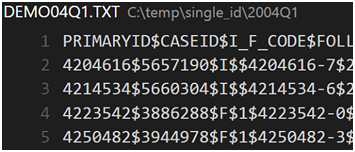
We process not only FAERS data files but also the older AERS data. All records in these files are delimitated by newline (\n), and attributes by dollar sign ($), as illustrated in the following example snapshot. However, we found two peculiar cases that need special care, i.e., newline character in a record and abnormal attribute delimiter.
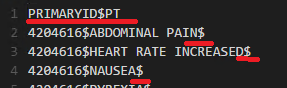
Some records contain newline characters (\n), like drug name or PT value. We guess FAERS did not examine every end-user's report. This character would wrongly divide a single record into two rows. As such, we checked each record to make sure the correct number of attributes, and resolved those with incorrect newline characters.
According to FAERS data format, the first row defines the names of attributes. Consider the following example, which contains two attributes (PRIMARYID and PT), so every row should have two values delimitated by only one delimiter (
FAERS attributes have been changed several times, which are summarized in the following table. An empty cell represents absence of the attribute in that time. For example, CASEVERSION was not introduced till 2012Q4, so it is absent before that time.
This summarization also indicates the evolution of attribute names, which are highlighted with red color. For example, CASEID has been changed from CASE since 12Q3 and PRIMARYID changed from ISR.
For those attributes that are still “active” in the current release, we always adopt the newest name, disregarding the time the attribute is introduced or changed, while for those “inactive” attributes, i.e., they are no longer used by FAERS, we still keep them but filled with missing values.
Another noticed issue is some attribute names are collided with SQL keywords. We append a baseline to distinguish them from SQL keywords. For example, the "ROUTE" attribute in the DRUG table is replaced by "ROUTE_":
You can find a database's meta data in scripts/metadata.py
We introduced several new attributes in the DEMO and DRUG tables.
-
DEMO (DEMOGRAPHIC)
-
-WT_KG: This weight attribute is calculated from WT and WT_COD, with unit in KG
-
-AGE_TYPE: A discretization of AGE and AGE_COD attribute into 10 tags, based on the Age group in MeSH. You can check detail in this table.
-
-
DRUG
- RXCUI: We transform the DRUGNAME attribute into rxcui code.
Each quarter directory contains 7 files, including DEMO (DEMOGRAPHIC), DRUG (DRUG), REAC (REACTION), OUTC (OUTCOME), RPSR (REPORT), THER (THERAPY), and INDI (INDICATIONS). For example,
.\data
|>2004Q1
|>DEMO04Q1.TXT
|>DRUG04Q1.TXT
|> ...
|>2004Q2
|>2004Q3
|> ...