-
Notifications
You must be signed in to change notification settings - Fork 4
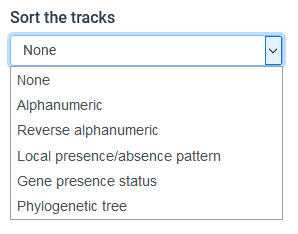
Sorting options
As of v1.0.0 Panache offers multiple sorting options for the genomes in the Presence / Absence Matrix. All of them can be chosen through the "Sort the tracks" button, but may require additional files to be loaded.
Please find below their effects and recquirements.
- None
- Alphanumerical
- Reverse Alphanumerical
- Phylogenetic tree
- Gene presence/absence search
- Local PAV pattern
Sorting option that returns genomes in their original order of appearance within the pangenome file.
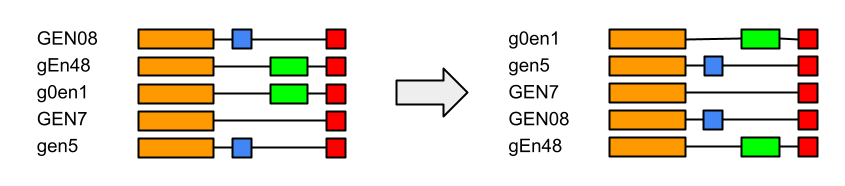
Offers an alphanumerical sort following certain criterias:
- Case insensitive
- Numbers first
- Numeric sequences count as integers, ordered increasingly
Below is an exemple of these criterias in action:
Same as the Alphanumerical option, but reversed.
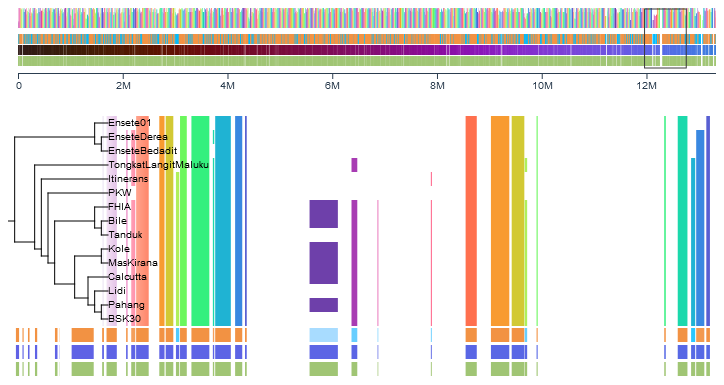
Only available when a newick file (.nwk) is uploaded.
This option will order the genomes according to their positions in the provided hierarchical tree, following the pre-established clusters.
The phylogenetic tree can be toggled on and off thanks to the "Display phylogenetic tree" button. Please note that it has to be toggled off if you wish to use another sorting order.
DISCLAIMER: for now, this sorting options only works when the pangenome file is built with the same genes that are annotated in the gff file. Further work will extend this sorting option to cases were blocks and annotations do not share their coordinates.
Only available when a gff3 file is uploaded.
When selected, a search bar si made available through the "Choose gene" buttonn, where the user can input the ID of a gene of their choice from anywhere in the pangenome. A tag will be created for the gene, and the user can then toggle its desired status, between presence and absence.

Once all tags are choosen, the sort button will consider the tags as criterias and calculate a matching score for each genome. Genomes will be ordered from top to bottom based on their matching scores, best first.
The scores are available within a tooltip by hovering the "Information about search" button once the sorting is completed.

Orders genomes with a Hierarchical Agglomerative Clustering (HAC) based on presence / absence motifs in a selected region. When chosen, a slider appears on screen and can be used to delimit the region for the calculation of the hierarchy.* Genomes will be grouped based on their similarities of succession of presence and absence within the selected region, and ordered accordingly on screen.
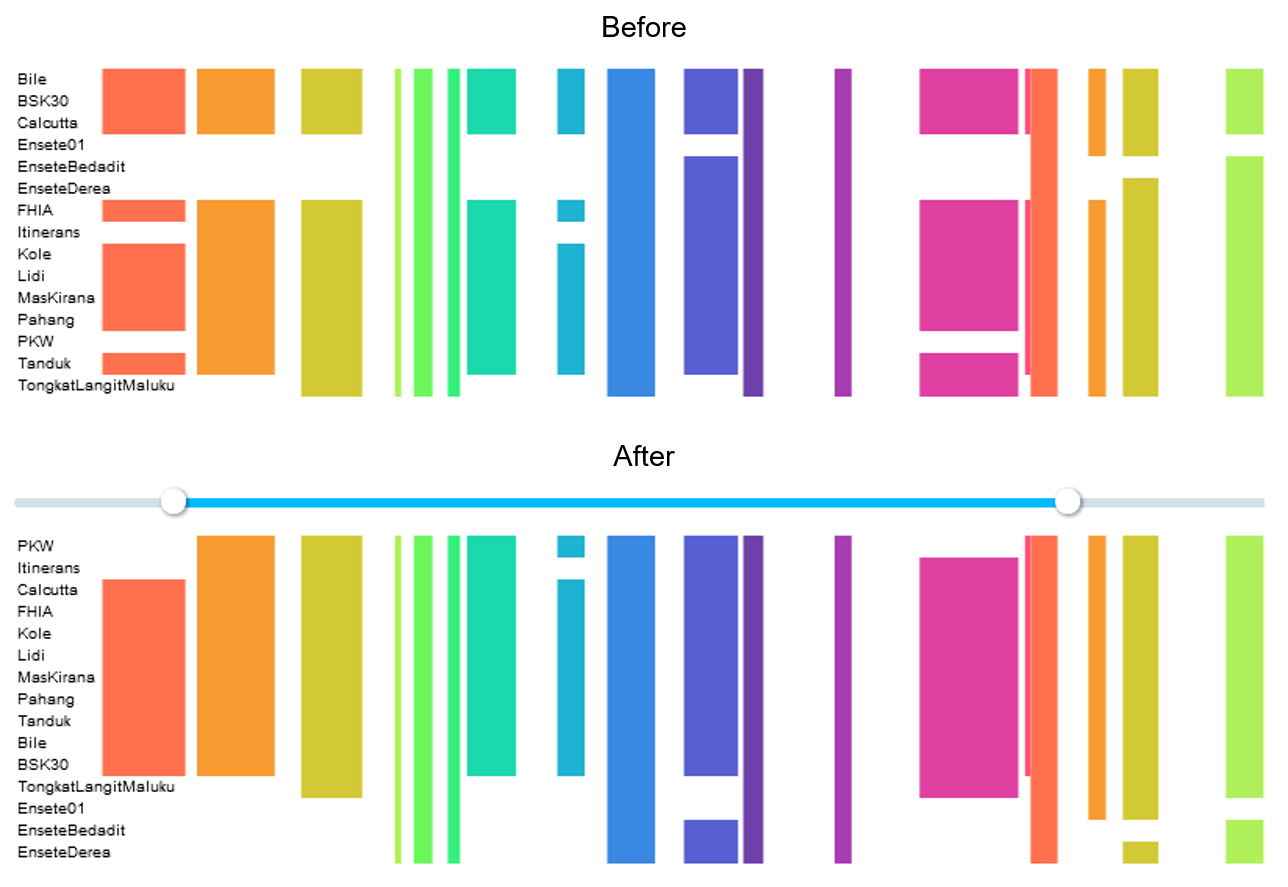
CAUTION: Only the blocks that are fully within the chosen range will be considered when clustering.
Our HAC algorithm, implemented with the library greenelab/hclust, uses a Simple Matching coefficient (which gives the same weight to matching absences than to matching presences) for the calculation of the dissimilarity matrix, and UPGMA - Unweighted Pair-Groups Method Average for the clustering. More about hierarchical clustering here, with helpful explanation and examples.