-
Notifications
You must be signed in to change notification settings - Fork 30
openmalariaTools
Note: This software is unmaintained and not related to the openmalaria.tools repository
A GUI frontend for managing, creating and running local experiments.
OpenMalariaTools is a graphical user interface created for an easier interaction with the OpenMalaria scientific application.
OpenMalariaTools is developed as a cross-platform application using Python and PyGTK. For some of its features, OpenMalariaTools requires a Java run-time environment.
OpenMalariaTools is currently in beta stage 6. It runs on both Linux and Windows, but currently not on the Mac OS due to the unavailability of PyGTK natively on the Mac.
The beta 6 release uses schema version 29 (beta) and has been packaged with an installer for windows and debian packages for linux. To use:
- Make sure a Java run-time environment is installed.
- Install Python (version 2.6 or 2.7; the 3.x series won't work) and PyGTK. (The OpenMalariaTools installer should install PyGTK, but it does not install Python itself)
- Download and install one of the following:
- (Windows installer) OpenMalariaTools_installer_v00.08.exe
- (linux 32-bit .deb) OpenMalariaTools_00.08_i386.deb
Alternatively, one can install from source (familiarity with subversion or TortoiseSVN assumed).
- Make sure a Java run-time environment, python and PyGTK are installed (as above).
- Check out the source code for OpenMalariaTools:
svn checkout http://openmalaria.googlecode.com/svn/application_deployment OpenMalariaTools
- If needed, replace (update) the
application/openMalaria[.exe]binary.
OpenMalariaTools features:
- Experiment creator
- Basic scenario editor
- Batch mode
- LiveGraph© integration
- Automatic schema translation
- Custom population size setting (for test batches)
To run some scenarios:
- Open the application
- Click on the load button
- Select some scenario files. If the input scenario files were written to suit a legacy schema version, the tool will enquire whether it should attempt to automatically update the scenario files to the current schema. It could happen that an automatic translation isn't possible, in which case you will need to update the outdated scenario manually.
- Add options to a model run or select a specific output folder.
* Once a scenario is selected, the LiveGraph
©option is enabled. Thus option allows you to start a third party software that will plot the data as it is produced by the OpenMalaria simulator. Be aware that LiveGraph©will look for specific parameters in the scenario file's monitoring section. * Single output folder. Instead of having one output folder per scenario file, you may opt to have one output folder for all the selected scenarios. This is useful if you're running many scenarios (e.g., in batch mode). * Custom population size. The population size can be customized for all selected scenarios. This feature is particularly useful if you want to test a scenario with a smaller population size before committing computer resources for a longer simulation. * If you are running OpenMalaria on a Windows 7 or Vista platform, the operating systems' security settings may prevent you from writing data to the default output folder. You should then select another path (e.g., your home folder). - When you have completed the simulations set up, click on the start button under the terminal screen and wait. You may follow your scenarios' progression on the terminal or review the situation summary in scenarios' list where you can see the status of each simulation (success, failure, or not yet started).
| overview |
|---|
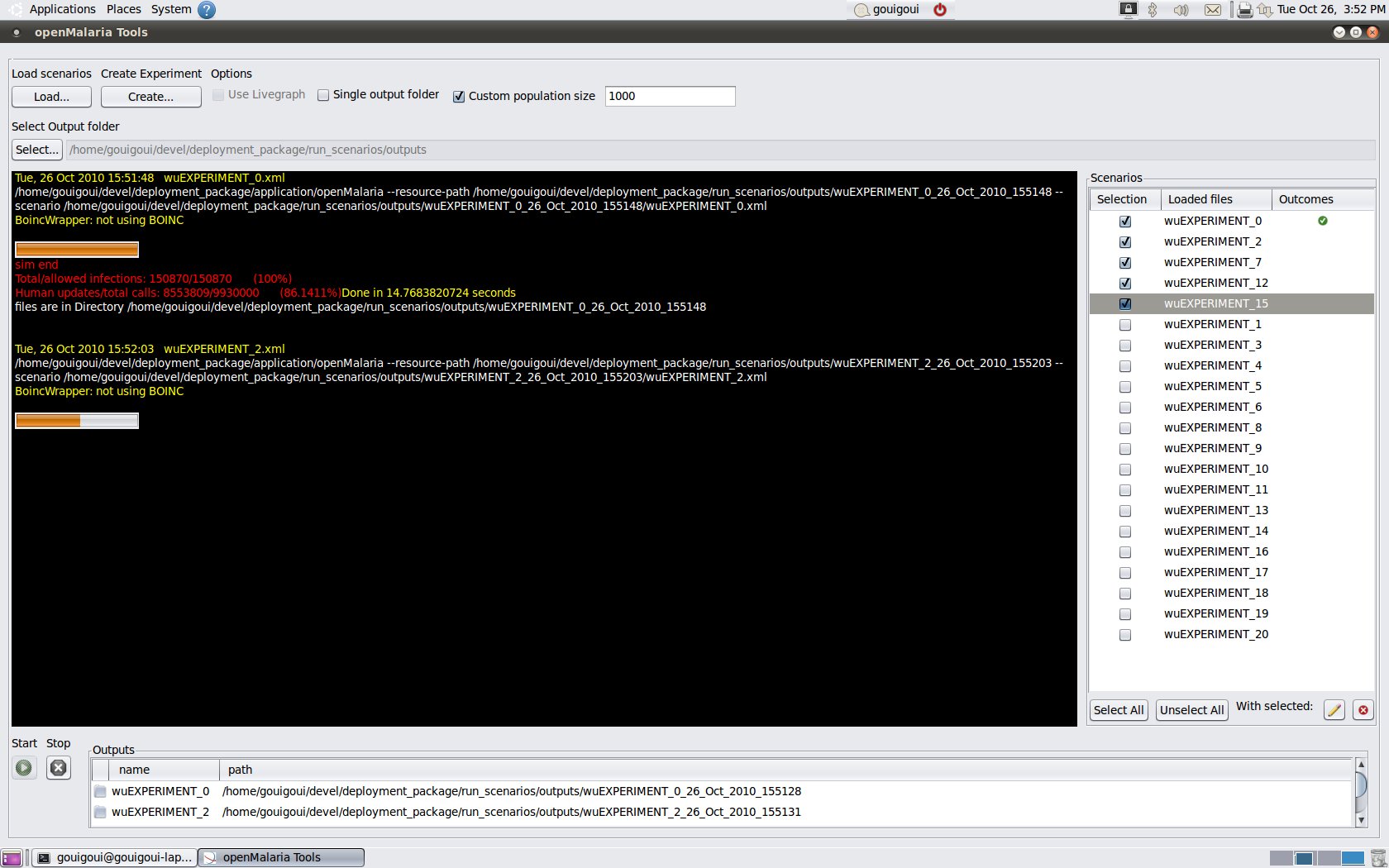 |
| livegraph |
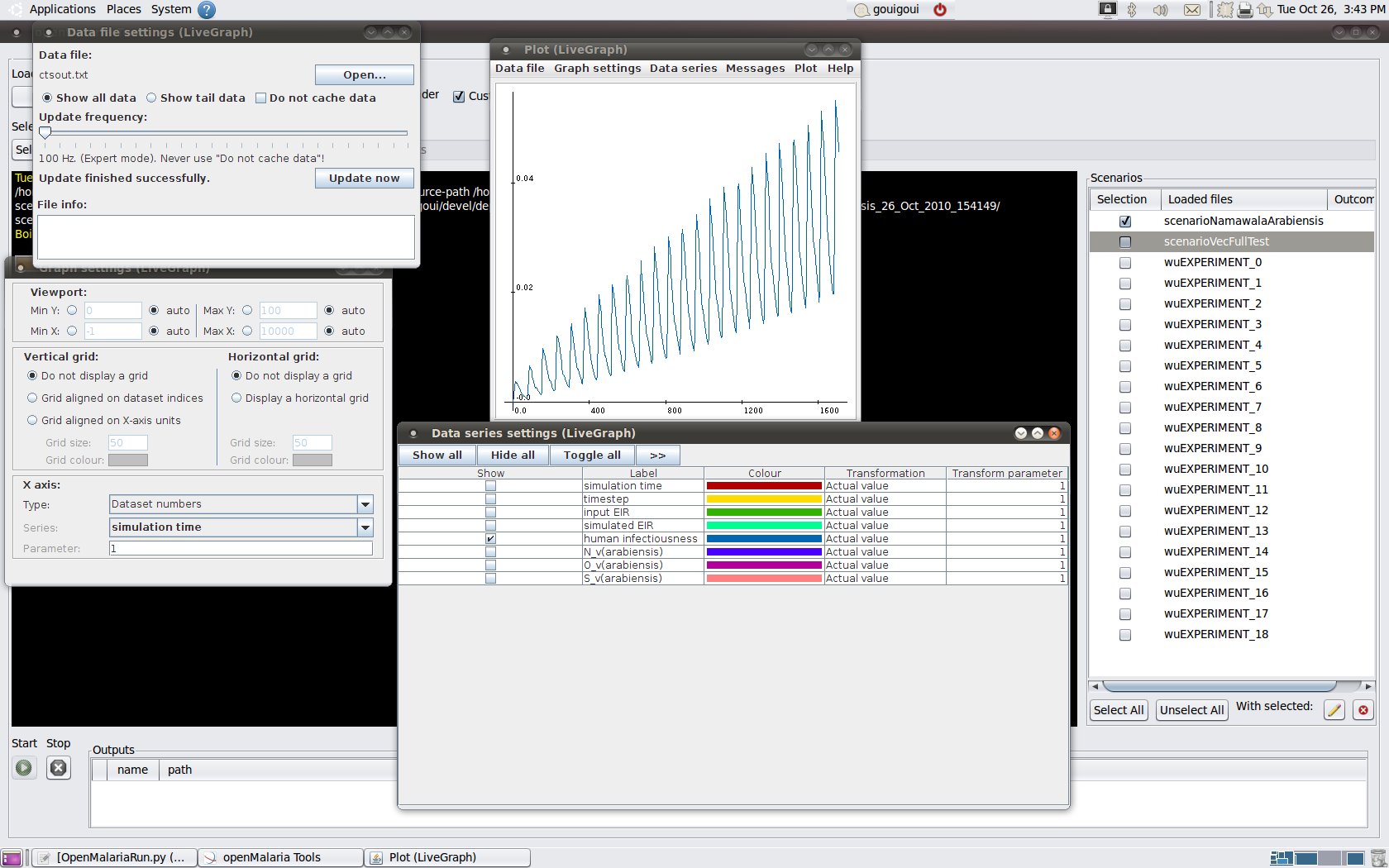 |
| editor |
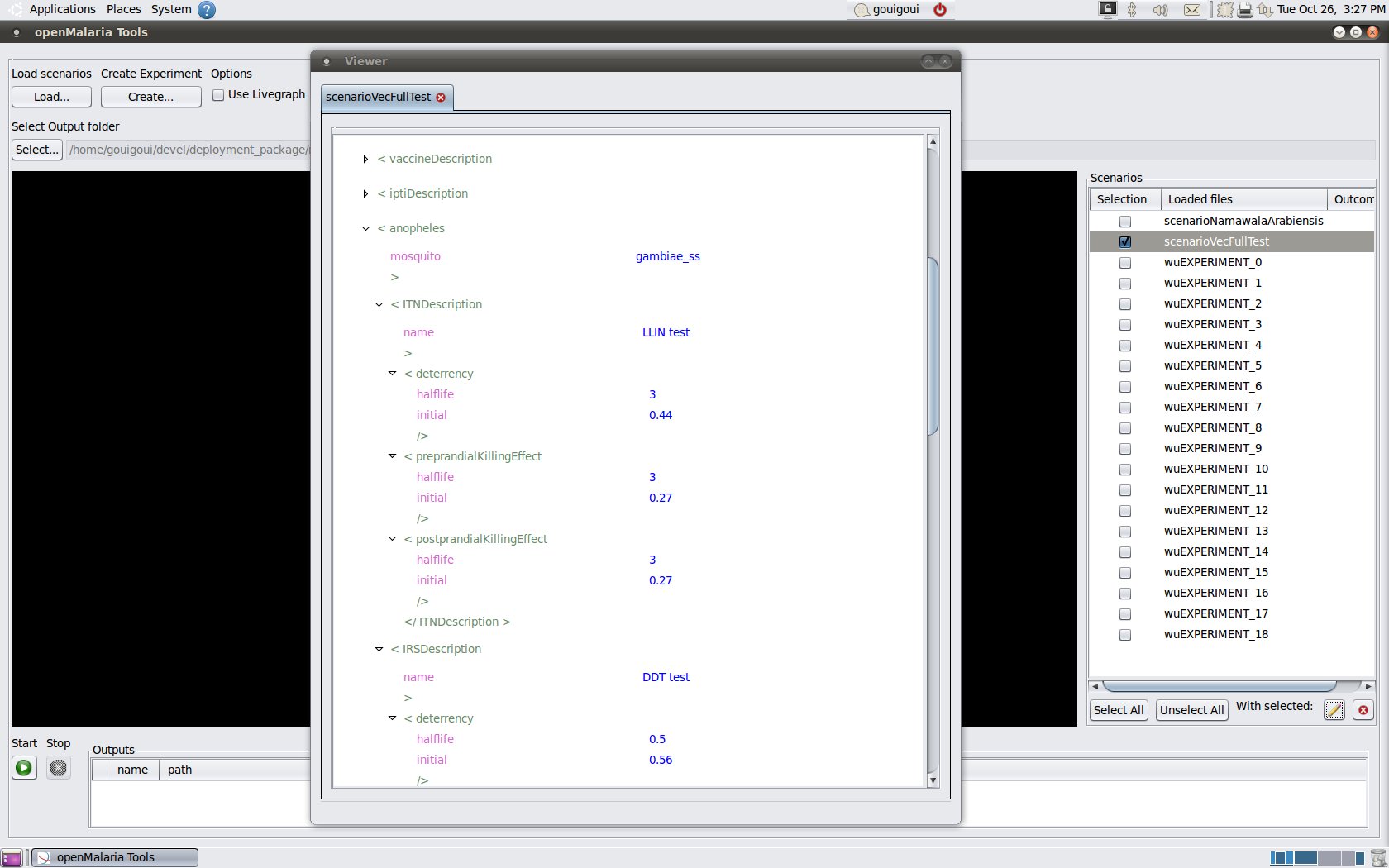 |
| experiment creator |
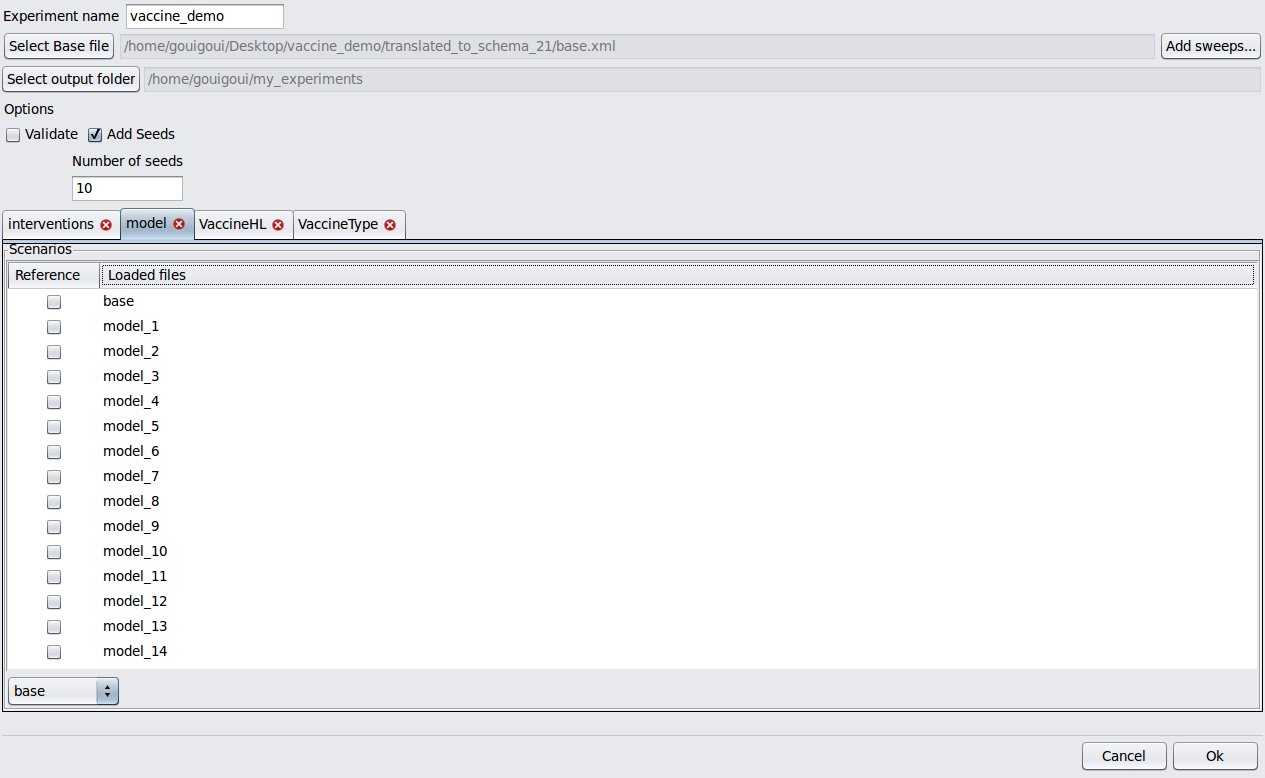 |
| Download openmalaria | Installation instructions | XML Schema Documentation |
| XML Schema Version | Program version | master |
develop |
|---|---|---|---|
| 43 | schema-43.0 |
|
|
- User Guide
- Compilation Guide
- Developer Guide
- Schema Update Guide
- Scenario Design Guide
- Monitoring Guide
- Changelog
- Schema Documentation
- Human demography
- Levels of transmission
- Parasite dynamics within humans
- P vivax dynamics
- Vector bionomics and transmission to humans
- Mosquito population dynamics
- Clinical (illness) models
- Time in the models