This repository is a container of methods that Neurodata usees to expose their open-source code while it is in the process of being merged with larger scientific libraries such as scipy, scikit-image, or scikit-learn. Additionally, methods for computational neuroscience on brains too specific for a general scientific library can be found here, such as image registration software tuned specifically for large brain volumes.
The repository originated as the project of a team in Joshua Vogelstein's class Neurodata at Johns Hopkins University. This project was focused on data science towards the mouselight data. It becme apparent that the tools developed for the class would be useful for other groups doing data science on large data volumes. The repository can now be considered a "holding bay" for code developed by Neurodata for collaborators and researchers to use.
- get conda
- create a virtual environment:
conda create --name brainlit python=3.8 - activate the environment:
conda activate brainlit
- install brainlit:
pip install brainlit
- clone the repo:
git clone https://github.com/neurodata/brainlit.git - cd into the repo:
cd brainlit - install brainlit:
pip install -e .
The source data directory should have an octree data structure
data/
├── default.0.tif
├── transform.txt
├── 1/
│ ├── 1/, ..., 8/
│ └── default.0.tif
├── 2/ ... 8/
└── consensus-swcs (optional)
├── G-001.swc
├── G-002.swc
└── default.0.tif
If your team wants to interact with cloud data, each member will need account credentials specified in ~/.cloudvolume/secrets/x-secret.json, where x is one of [aws, gc, azure] which contains your id and secret key for your cloud platform.
We provide a template for aws in the repo for convenience.
Each user will start their scripts with approximately the same lines:
from brainlit.utils.ngl import NeuroglancerSession
session = NeuroglancerSession(url='file:///abc123xyz')
From here, any number of tools can be run such as the visualization or annotation tools. Interactive demo.
The registration subpackage is a facsimile of ARDENT, a pip-installable (pip install ardent) package for nonlinear image registration wrapped in an object-oriented framework for ease of use. This is an implementation of the LDDMM algorithm with modifications, written by Devin Crowley and based on "Diffeomorphic registration with intensity transformation and missing data: Application to 3D digital pathology of Alzheimer's disease." This paper extends on an older LDDMM paper, "Computing large deformation metric mappings via geodesic flows of diffeomorphisms."
This is the more recent paper:
Tward, Daniel, et al. "Diffeomorphic registration with intensity transformation and missing data: Application to 3D digital pathology of Alzheimer's disease." Frontiers in neuroscience 14 (2020).
https://doi.org/10.3389/fnins.2020.00052
This is the original LDDMM paper:
Beg, M. Faisal, et al. "Computing large deformation metric mappings via geodesic flows of diffeomorphisms." International journal of computer vision 61.2 (2005): 139-157.
https://doi.org/10.1023/B:VISI.0000043755.93987.aa
A tutorial is available in docs/notebooks/registration_demo.ipynb.
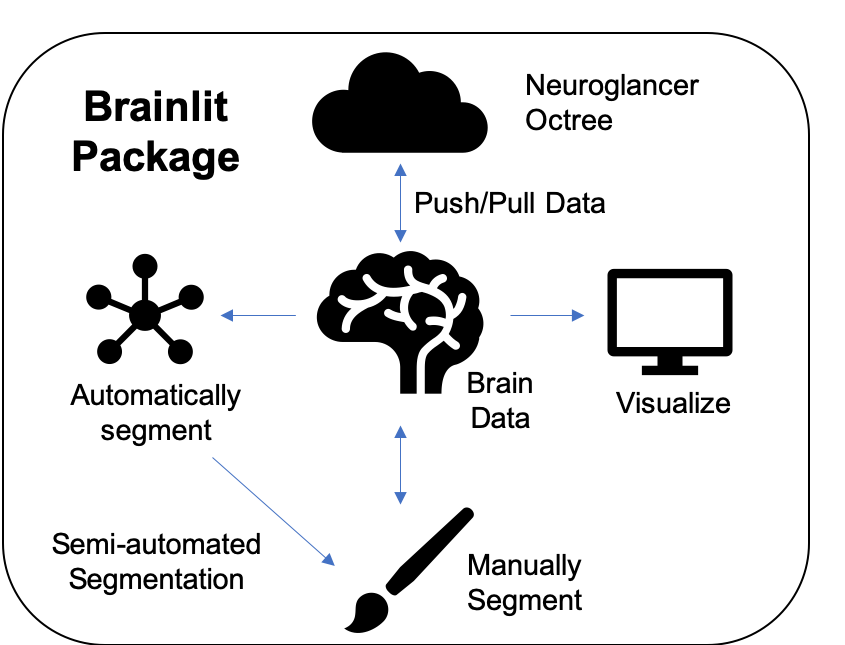
The core brain-lit package can be described by the diagram at the top of the readme:
Brainlit uses the Seung Lab's Cloudvolume package to push and pull data through the cloud or a local machine in an efficient and parallelized fashion. Interactive demo.
The only requirement is to have an account on a cloud service on s3, azure, or google cloud.
Loading data via local filepath of an octree structure is also supported. Interactive demo.
Brainlit supports many methods to visualize large data. Visualizing the entire data can be done via Google's Neuroglancer, which provides a web link as shown below.
screenshot
Brainlit also has tools to visualize chunks of data as 2d slices or as a 3d model. Interactive demo.
screenshot
Brainlit includes a lightweight manual segmentation pipeline. This allows collaborators of a projec to pull data from the cloud, create annotations, and push their annotations back up as a separate channel. Interactive demo.
Similar to the above pipeline, segmentations can be automatically or semi-automatically generated and pushed to a separate channel for viewing. Interactive demo.
The documentation can be found at https://brainlight.readthedocs.io/en/latest/.
Running tests can easily be done by moving to the root directory of the brainlit package ant typing pytest tests or python -m pytest tests.
Running a specific test, such as test_upload.py can be done simply by ptest tests/test_upload.py.
Contribution guidelines can be found via CONTRIBUTING.md
Thanks to the neurodata team and the group in the neurodata class which started the project. This project is currently managed by Tommy Athey and Bijan Varjavand.