Starting from v0.3.0, this package will not be updated, bugfixes will still be included if issues are raised. You can already find RDST in the Aeon package at https://github.com/aeon-toolkit/ . Further improvements are planned for further speeding up RDST, these improvement will only be implemented in aeon.
ALL FUNCTIONALITIES OF THIS PACKAGE OUTSIDE OF THE INTEPRETER ARE NOW PORTED INTO AEON FROM V0.6.0, PLEASE REFER TO THE AEON IMPLEMENTATION WHEN DOING EXPERIMENTS.
AN EXAMPLE NOTEBOOK ON HOW TO CORRECTLY INTERPRET SHAPELETS FROM RDST IS PLANNED (see aeon-toolkit/aeon#973)
If these functionnalities are what you need, I highly recommend that you use aeon as I spent more time on the aeon implementation and tests compared to convst.
Welcome to the convst repository. It contains the implementation of the Random Dilated Shapelet Transform (RDST) along with other works in the same area.
This work was supported by the following organisations:
| Overview | |
|---|---|
| Compatibility |  |
| CI/CD |   |
| Code Quality |  |
| Downloads |
The recommended way to install the latest stable version is to use pip with pip install convst. To install the package from sources, you can download the latest version on GitHub and run python setup.py install. This should install the package and automatically look for the dependencies using pip.
We recommend doing this in a new virtual environment using anaconda to avoid any conflict with an existing installation. If you wish to install dependencies individually, you can see dependencies in the setup.py file.
An optional dependency that can help speed up numba, which is used in our implementation, is the Intel vector math library (SVML). When using conda it can be installed by running conda install -c numba icc_rt. I didn't test the behavior with AMD processors, but I suspect it won't work.
We give here a minimal example to run the RDST algorithm on any dataset of the UCR archive using the aeon API to get datasets:
from convst.classifiers import R_DST_Ridge
from convst.utils.dataset_utils import load_UCR_UEA_dataset_split
X_train, X_test, y_train, y_test, _ = load_UCR_UEA_dataset_split('GunPoint')
# First run may be slow due to numba compilations on the first call.
# Run a small dataset like GunPoint if this is the first time you call RDST on your system.
# You can change n_shapelets to 1 to make this process faster. The n_jobs parameter can
# also be changed to increase speed once numba compilation are done.
rdst = R_DST_Ridge(n_shapelets=10_000, n_jobs=1).fit(X_train, y_train)
print("Accuracy Score for RDST : {}".format(rdst.score(X_test, y_test)))If you want a more powerful model, you can use R_DST_Ensemble as follows (note that additional Numba compilation might be needed here):
from convst.classifiers import R_DST_Ensemble
rdst_e = R_DST_Ensemble(
n_shapelets_per_estimator=10_000,
n_jobs=1
).fit(X_train, y_train)
print("Accuracy Score for RDST : {}".format(rdst_e.score(X_test, y_test)))You can obtain faster result by using more jobs and even faster, at the expense of some accuracy, with the prime_dilation option:
rdst_e = R_DST_Ensemble(
n_shapelets_per_estimator=10_000,
prime_dilations=True,
n_jobs=-1
).fit(X_train, y_train)
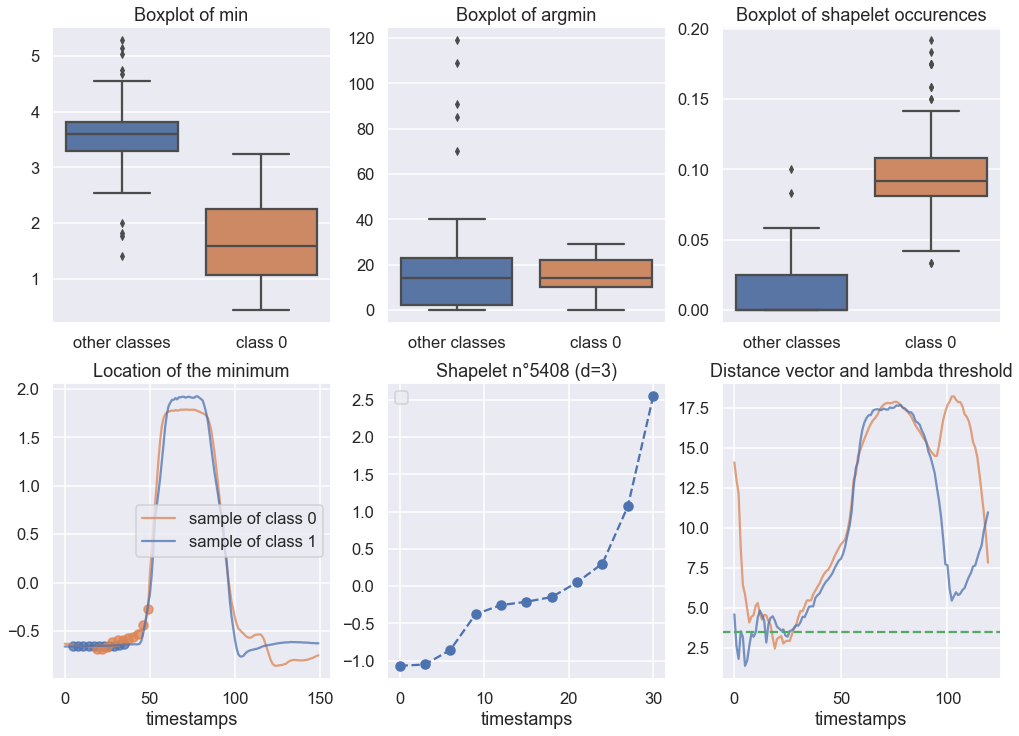
print("Accuracy Score for RDST : {}".format(rdst_e.score(X_test, y_test)))You can also visualize a shapelet using the visualization tool to obtain such visualization :
To know more about all the interpretability tools, check the documentation on readthedocs.
RDST support the following type of time series:
- Univariate and same length
- Univariate and variable length
- Multivariate and same length
- Multivariate and variable length
We use the standard scikit-learn interface and expect as input a 3D numpy array of shape (n_samples, n_features, n_timestamps). For variable length input, we expect a (python) list of numpy arrays, or a numpy array with object dtype.
Multiple scripts are available under the PaperScripts folder. It contains the exact same scripts used to generate our results, notably the test_models.py file, used to generate the csv results available in the Results folder of the archive.
If you are experiencing bugs in the RDST implementation, or would like to contribute in any way, please create an issue or pull request in this repository. For other question or to take contact with me, you can email me at antoine.guillaume45@gmail.com
If you use our algorithm or publication in any work, please cite the following paper (ArXiv version https://arxiv.org/abs/2109.13514):
@InProceedings{10.1007/978-3-031-09037-0_53,
author="Guillaume, Antoine
and Vrain, Christel
and Elloumi, Wael",
title="Random Dilated Shapelet Transform: A New Approach for Time Series Shapelets",
booktitle="Pattern Recognition and Artificial Intelligence",
year="2022",
publisher="Springer International Publishing",
address="Cham",
pages="653--664",
abstract="Shapelet-based algorithms are widely used for time series classification because of their ease of interpretation, but they are currently outperformed by recent state-of-the-art approaches. We present a new formulation of time series shapelets including the notion of dilation, and we introduce a new shapelet feature to enhance their discriminative power for classification. Experiments performed on 112 datasets show that our method improves on the state-of-the-art shapelet algorithm, and achieves comparable accuracy to recent state-of-the-art approaches, without sacrificing neither scalability, nor interpretability.",
isbn="978-3-031-09037-0"
}To cite the RDST Ensemble method, you can cite the PhD thesis where it is presented as (soon to be available, citation format may change):
@phdthesis{Guillaume2023,
author="Guillaume, Antoine",
title="Time series classification with Shapelets: Application to predictive maintenance on event logs",
school="University of Orléans",
year="2023",
url="https://www.theses.fr/s265104"
}- Finish Numpy docs in all python files
- Update documentation and examples
- Enhance interface for interpretability tools
- Add the Generalised version of RDST
- Continue unit tests and code coverage/quality
Here are the code-related citations that were not made in the paper
[2]: The Numpy development team, "Array programming with NumPy", Nature 2020


.png)