-
Notifications
You must be signed in to change notification settings - Fork 0
BioRubyOnRails
This page describes how to use BioRuby with your Rails application.
BioRuby comes with an interactive shell which can be used in a console or in a web browser as a Rails plugin.
Installing the BioRuby 1.3.0 release as a Ruby gems package.
% sudo gem install bio
Password:
Successfully installed bio-1.3.0
1 gem installed
Installing ri documentation for bio-1.3.0...
Installing RDoc documentation for bio-1.3.0...
If your gem stick at the following message
Bulk updating Gem source index for: http://gems.rubyforge.org
and took very long time to find the BioRuby gem, you'd better to update the RubyGems itself.
% gem -v
1.0.1
% sudo gem update --system
Updating RubyGems...
Bulk updating Gem source index for: http://gems.rubyforge.org
Attempting remote update of rubygems-update
Successfully installed rubygems-update-1.3.1
1 gem installed
Updating version of RubyGems to 1.3.1
Installing RubyGems 1.3.1
:
RubyGems system software updated
% gem -v
1.3.1
OK. Test the BioRuby shell in a console.
% bioruby
Loading config (/Users/xxx/.bioruby/shell/session/config) ... done
Loading object (/Users/xxx/.bioruby/shell/session/object) ... done
Loading history (/Users/xxx/.bioruby/shell/session/history) ... done
. . . B i o R u b y i n t h e s h e l l . . .
Version : BioRuby 1.3.0 / Ruby 1.8.6
bioruby> seq = getseq("atgc" * 10)
==> #<Bio::Sequence:0x117de40 @seq="atgcatgcatgcatgcatgcatgcatgcatgcatgcatgc", @moltype=Bio::Sequence::NA>
bioruby> seq.randomize
==> "ttcgagaacaccgcgatcgttgcaagattcgttatcaggc"
bioruby> seq.randomize.translate
==> "HFGLYRN*RGQRL"
bioruby> exit
. . . B i o R u b y i n t h e s h e l l . . .
Saving history (/Users/full/.bioruby/shell/session/history) ... done
Saving object (/Users/full/.bioruby/shell/session/object) ... done
Saving config (/Users/full/.bioruby/shell/session/config) ... done
Create an Rails project (you can also use existing Rails application as well).
% rails -v
Rails 1.2.6
% rails hoge
create
create app/controllers
:
create log/test.log
Activate the BioRuby shell as a Rails plugin.
% bioruby --rails hoge
Creating directory (/Users/ktym/hoge/shell/session) ... done
Creating directory (/Users/ktym/hoge/shell/plugin) ... done
Creating directory (/Users/ktym/hoge/data) ... done
Installing Rails plugin for BioRuby shell ...
exists app/controllers
exists app/helpers
create app/views/bioruby
exists app/views/layouts
create public/images/bioruby
exists public/stylesheets
create app/controllers/bioruby_controller.rb
create app/helpers/bioruby_helper.rb
create app/views/bioruby/_methods.rhtml
create app/views/bioruby/_classes.rhtml
create app/views/bioruby/_modules.rhtml
create app/views/bioruby/_log.rhtml
create app/views/bioruby/_variables.rhtml
create app/views/bioruby/commands.rhtml
create app/views/bioruby/history.rhtml
create app/views/bioruby/index.rhtml
create app/views/layouts/bioruby.rhtml
create public/images/bioruby/spinner.gif
create public/images/bioruby/gem.png
create public/images/bioruby/link.gif
create public/images/bioruby/bg.gif
create public/stylesheets/bioruby.css
done
>>>
>>> open http://localhost:3000/bioruby
>>>
(You can change the port number by adding "-- -p 4000" option)
. . . B i o R u b y i n t h e s h e l l . . .
Version : BioRuby 1.3.0 / Ruby 1.8.7
=> Booting WEBrick...
=> Rails application started on http://0.0.0.0:3000
=> Ctrl-C to shutdown server; call with --help for options
[2009-02-25 01:19:29] INFO WEBrick 1.3.1
[2009-02-25 01:19:29] INFO ruby 1.8.7 (2008-06-09) [i686-darwin9.3.0]
[2009-02-25 01:19:29] INFO WEBrick::HTTPServer#start: pid=11036 port=3000
You may see the different message as follows if you have installed a Mongrel web server for Rails.
. . . B i o R u b y i n t h e s h e l l . . .
Version : BioRuby 1.3.0 / Ruby 1.8.6
./config/boot.rb:20:Warning: Gem::SourceIndex#search support for String patterns is deprecated
=> Booting Mongrel (use 'script/server webrick' to force WEBrick)
=> Rails application starting on http://0.0.0.0:3000
=> Call with -d to detach
=> Ctrl-C to shutdown server
/Library/Ruby/Gems/1.8/gems/mongrel-1.1.5/bin/mongrel_rails:60: warning: conflicting chdir during another chdir block
** Starting Mongrel listening at 0.0.0.0:3000
** Starting Rails with development environment...
** Rails loaded.
** Loading any Rails specific GemPlugins
** Signals ready. TERM => stop. USR2 => restart. INT => stop (no restart).
** Rails signals registered. HUP => reload (without restart). It might not work well.
** Mongrel 1.1.5 available at 0.0.0.0:3000
** Use CTRL-C to stop.
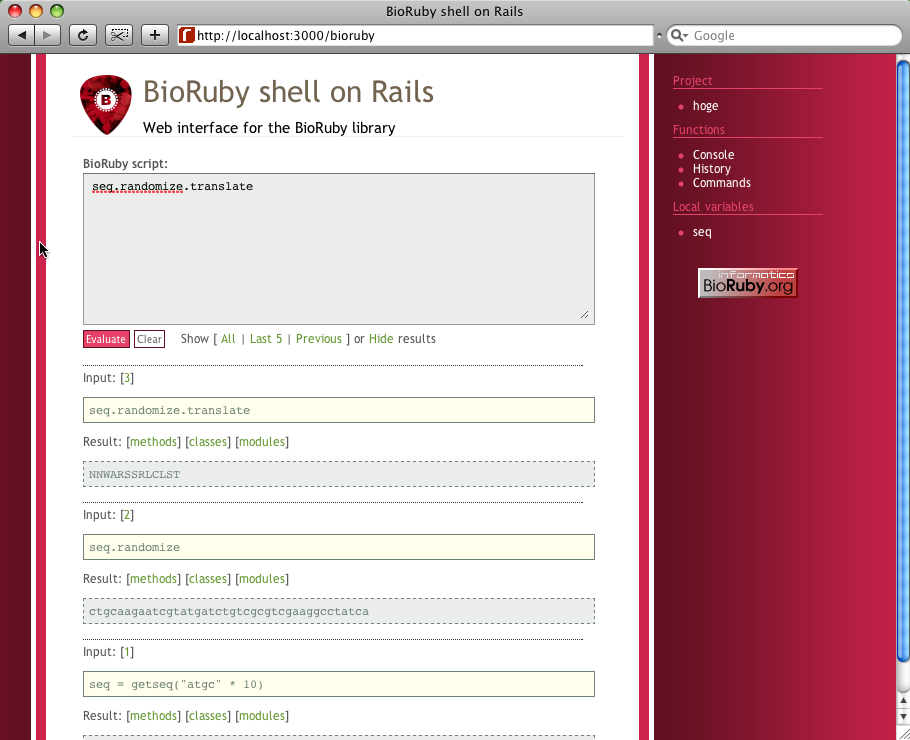
Anyway, that's it. You are ready to use the BioRuby shell on Rails now.
Open the following URL in your web browser.
http://localhost:3000/bioruby
You can now able to use all of Ruby and BioRuby functionalities even with your Rails models in your browser. Enjoy!