Are you using the tidyverse with an OMOP Common Data Model?
Interact with your CDM in a pipe-friendly way with CDMConnector.
- Quickly connect to your CDM and start exploring.
- Build data analysis pipelines using familiar dplyr verbs.
- Easily extract subsets of CDM data from a database.
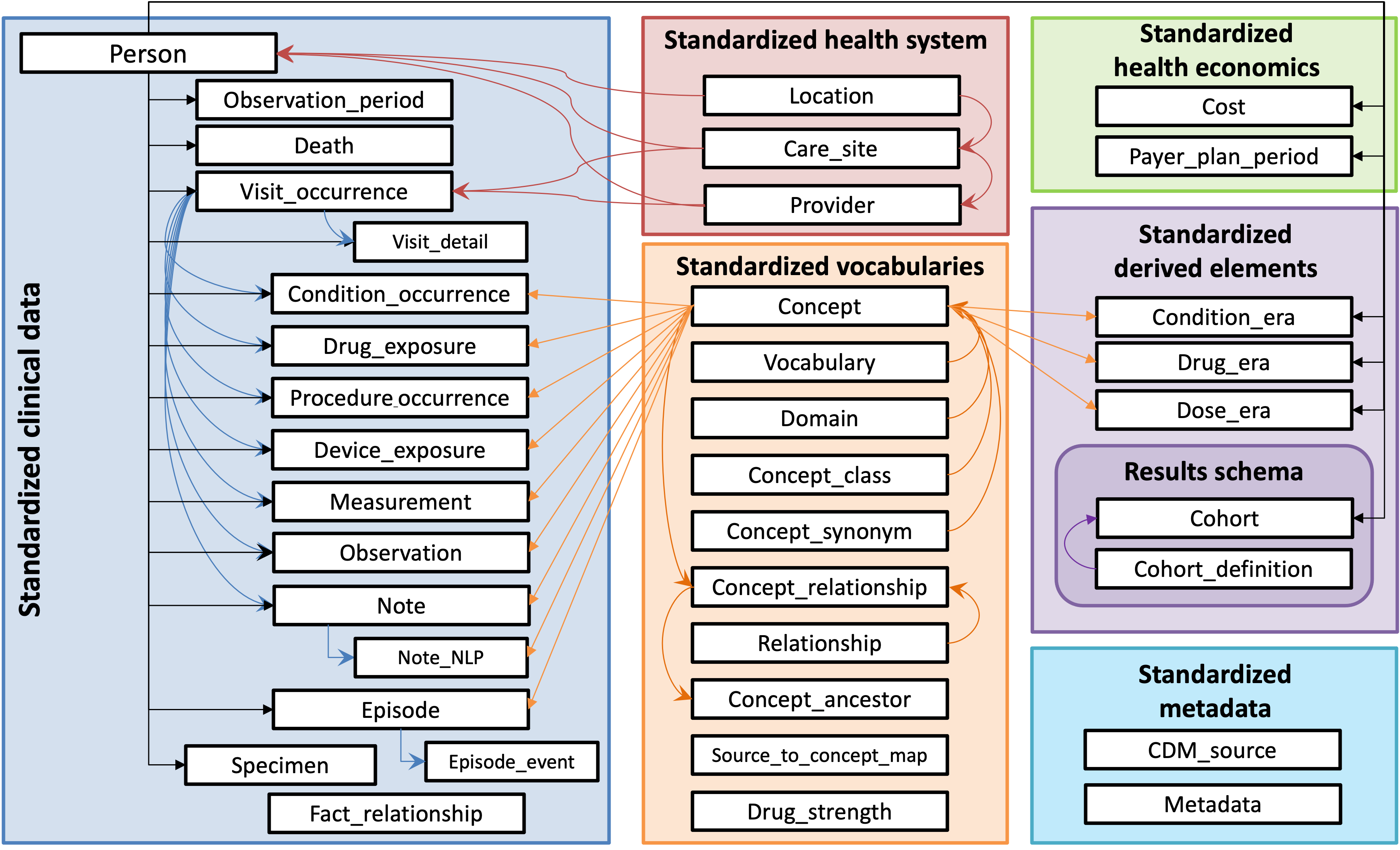
CDMConnector introduces a single R object that represents an OMOP CDM relational database inspired by the dm, DatabaseConnector, and Andromeda packages. The cdm objects encapsulate references to OMOP CDM tables in a remote RDBMS as well as metadata necessary for interacting with a CDM, allowing for dplyr style data analysis pipelines and interactive data exploration.
CDMConnector is meant to be the entry point for composable tidyverse
style data analysis operations on an OMOP CDM. A cdm_reference object
behaves like a named list of tables.
- Quickly create a list of references to a subset of CDM tables
- Store connection information for later use inside functions
- Use any DBI driver back-end with the OMOP CDM
See Getting started for more details.
CDMConnector can be installed from CRAN:
install.packages("CDMConnector")The development version can be installed from GitHub:
# install.packages("devtools")
devtools::install_github("darwin-eu/CDMConnector")Create a cdm reference from any DBI connection to a database containing OMOP CDM tables. Use the cdm_schema argument to point to a particular schema in your database that contains your OMOP CDM tables and the write_schema to specify the schema where results tables can be created, and use cdm_name to provide a name for the database.
library(CDMConnector)
con <- DBI::dbConnect(duckdb::duckdb(dbdir = eunomia_dir()))
cdm <- cdm_from_con(con = con,
cdm_schema = "main",
write_schema = "main",
cdm_name = "my_duckdb_database")## Note: method with signature 'DBIConnection#Id' chosen for function 'dbExistsTable',
## target signature 'duckdb_connection#Id'.
## "duckdb_connection#ANY" would also be valid
A cdm_reference is a named list of table references:
library(dplyr)
names(cdm)## [1] "person" "observation_period" "visit_occurrence"
## [4] "visit_detail" "condition_occurrence" "drug_exposure"
## [7] "procedure_occurrence" "device_exposure" "measurement"
## [10] "observation" "death" "note"
## [13] "note_nlp" "specimen" "fact_relationship"
## [16] "location" "care_site" "provider"
## [19] "payer_plan_period" "cost" "drug_era"
## [22] "dose_era" "condition_era" "metadata"
## [25] "cdm_source" "concept" "vocabulary"
## [28] "domain" "concept_class" "concept_relationship"
## [31] "relationship" "concept_synonym" "concept_ancestor"
## [34] "source_to_concept_map" "drug_strength"
Use dplyr verbs with the table references.
cdm$person %>%
tally()## # Source: SQL [1 x 1]
## # Database: DuckDB v1.1.2 [root@Darwin 23.1.0:R 4.3.3//private/var/folders/2j/8z0yfn1j69q8sxjc7vj9yhz40000gp/T/RtmpDw9JTb/fileeea2255bd10b.duckdb]
## n
## <dbl>
## 1 2694
Compose operations with the pipe.
cdm$condition_era %>%
left_join(cdm$concept, by = c("condition_concept_id" = "concept_id")) %>%
count(top_conditions = concept_name, sort = TRUE)## # Source: SQL [?? x 2]
## # Database: DuckDB v1.1.2 [root@Darwin 23.1.0:R 4.3.3//private/var/folders/2j/8z0yfn1j69q8sxjc7vj9yhz40000gp/T/RtmpDw9JTb/fileeea2255bd10b.duckdb]
## # Ordered by: desc(n)
## top_conditions n
## <chr> <dbl>
## 1 Viral sinusitis 17268
## 2 Acute viral pharyngitis 10217
## 3 Acute bronchitis 8184
## 4 Otitis media 3561
## 5 Osteoarthritis 2694
## 6 Streptococcal sore throat 2656
## 7 Sprain of ankle 1915
## 8 Concussion with no loss of consciousness 1013
## 9 Sinusitis 1001
## 10 Acute bacterial sinusitis 939
## # ℹ more rows
And much more besides. See vignettes for further explanations on how to create database connections, make a cdm reference, and start analysing your data.
If you encounter a clear bug, please file an issue with a minimal reproducible example on GitHub.
## To cite package 'CDMConnector' in publications use:
##
## Black A, Gorbachev A, Burn E, Catala Sabate M (????). _CDMConnector:
## Connect to an OMOP Common Data Model_. R package version 1.6.0,
## https://github.com/darwin-eu/CDMConnector,
## <https://darwin-eu.github.io/CDMConnector/>.
##
## A BibTeX entry for LaTeX users is
##
## @Manual{,
## title = {CDMConnector: Connect to an OMOP Common Data Model},
## author = {Adam Black and Artem Gorbachev and Edward Burn and Marti {Catala Sabate}},
## note = {R package version 1.6.0, https://github.com/darwin-eu/CDMConnector},
## url = {https://darwin-eu.github.io/CDMConnector/},
## }
License: Apache 2.0