-
Notifications
You must be signed in to change notification settings - Fork 24
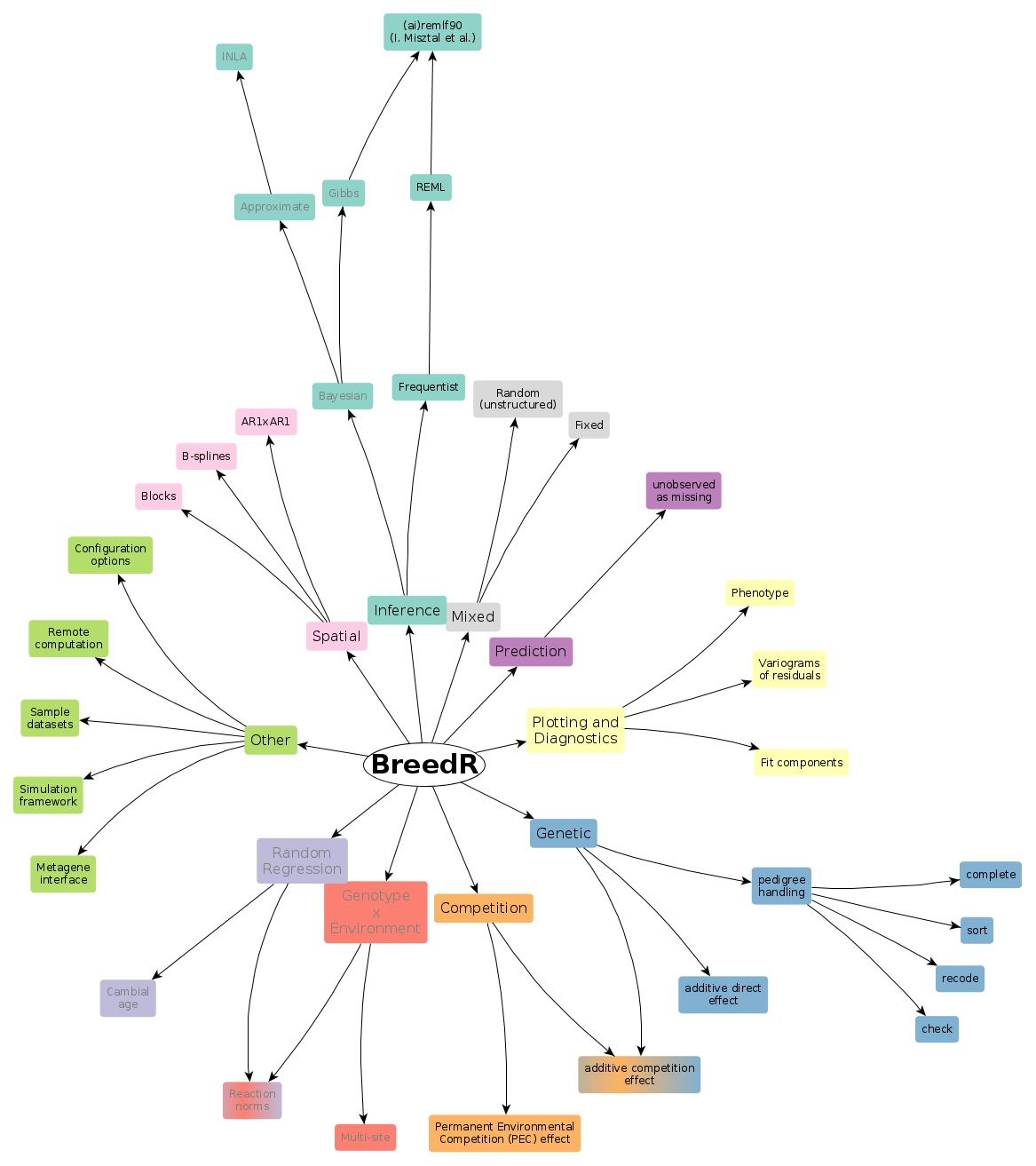
Handling pedigrees
Facundo Muñoz edited this page Apr 28, 2017
·
7 revisions
- What is a pedigree
- Checking pedigrees
- Building pedigrees
- Using a pedigree in an additive genetic effect
- Recovering Breeding Values in the original coding
- Recovering Breeding Values for the founders, in the original coding
- Identifying original codes from internal representation
-
A 3-column
data.frameormatrixwith the codes for each individual and its parents -
A family effect is easily translated into a pedigree:
- use the family code as the identification of a fictitious mother
- use
0orNAas codes for the unknown fathers
-
A pedigree sintetizes any kind of (genetic) relationship between individuals from one or more generations
| self | dad | mum |
|---|---|---|
| 69 | 0 | 64 |
| 70 | 0 | 41 |
| 71 | 0 | 56 |
| 72 | 0 | 55 |
| 73 | 0 | 22 |
| 74 | 0 | 50 |
- For computational reasons, the pedigree needs to meet certain conditions:
- Completness: all the individuals (also parents) must have an entry
- with possibly unknown parents (code
0orNA)
- with possibly unknown parents (code
- The offspring must follow the parents
- The codes must be sorted increasingly
- The codes must be consecutive
- Completness: all the individuals (also parents) must have an entry
- So, not every 3-column
data.frameormatrixwith codes is a proper pedigree:
set.seed(123); n.ped <- 5
ped.nightmare <- matrix(sample(30, n.ped*3), n.ped, 3,
dimnames = list(NULL, c('self', 'sire', 'dam')))
check_pedigree(ped.nightmare)## full_ped offsp_follows codes_sorted codes_consecutive
## FALSE FALSE FALSE FALSE
-
breedR implements a pedigree constructor that completes, sorts and recodes as necessary
-
The resulting object, of class
pedigreeis guranteed to meet the conditions
ped.fix <- build_pedigree(1:3, data = ped.nightmare)## Warning in build_pedigree(1:3, data = ped.nightmare): The pedigree has been
## recoded. Check attr(ped, 'map').
check_pedigree(ped.fix)## full_ped offsp_follows codes_sorted codes_consecutive
## TRUE TRUE TRUE TRUE
attr(ped.fix, 'map') # map from old to new codes## [1] NA 1 NA NA NA NA NA NA 3 4 NA 7 8 NA NA NA NA NA NA 2 5 6 10
## [24] 13 15 14 11 NA 12 9
| self | sire | dam |
|---|---|---|
| 9 | 2 | 20 |
| 23 | 13 | 30 |
| 12 | 21 | 22 |
| 24 | 27 | 29 |
| 25 | 10 | 26 |
| self | sire | dam |
|---|---|---|
| 1 | NA | NA |
| 2 | NA | NA |
| 3 | 1 | 2 |
| 4 | NA | NA |
| 5 | NA | NA |
| 6 | NA | NA |
| 7 | 5 | 6 |
| 8 | NA | NA |
| 9 | NA | NA |
| 10 | 8 | 9 |
| 11 | NA | NA |
| 12 | NA | NA |
| 13 | 11 | 12 |
| 14 | NA | NA |
| 15 | 4 | 14 |
- just include your original pedigree information and let breedR fix it for you
test.dat <- data.frame(ped.nightmare, y = rnorm(n.ped))
res.raw <- remlf90(fixed = y ~ 1,
genetic = list(model = 'add_animal',
pedigree = ped.nightmare,
# pedigree = test.dat[, 1:3], # same thing
var.ini = 1,
id = 'self'),
var.ini = list(resid = 1),
data = test.dat)## Warning in build_pedigree(1:3, data = ped.df): The pedigree has been
## recoded. Check attr(ped, 'map').
## pedigree has been recoded!
length(ranef(res.raw)$genetic)## [1] 15
## The pedigree used in the model matches the one manually built
identical(ped.fix, get_pedigree(res.raw))## [1] TRUE
## Predicted Breeding Valuess of the observed individuals
## Left-multiplying the vector of BLUP by the incidence matrix
## gives the BLUP of the observations in the right order.
Za <- model.matrix(res.raw)$genetic # incidence matrix
gen.blup <- with(ranef(res.raw),
cbind(value=genetic,
's.e.'=attr(genetic, 'se')))
PBVs <- Za %*% gen.blup
rownames(PBVs) <- test.dat$self| value | s.e. | |
|---|---|---|
| 9 | 0.21 | 0.89 |
| 23 | -1.29 | 0.89 |
| 12 | 0.42 | 0.89 |
| 24 | -0.18 | 0.89 |
| 25 | 0.84 | 0.89 |
## original codes of non-observed parents
(founders.orig <- setdiff(
sort(unique(as.vector(ped.nightmare[, c("sire", "dam")]))),
ped.nightmare[, "self"]
))## [1] 2 10 13 20 21 22 26 27 29 30
## map from original to internal codes
map.codes <- attr(get_pedigree(res.raw), "map")
## internal codes of non-observed parents
founders.int <- map.codes[founders.orig]
## Breeding Values of non-observed parents
founders.PBVs <- gen.blup[founders.int, ]
rownames(founders.PBVs) <- founders.orig| value | s.e. | |
|---|---|---|
| 2 | 0.11 | 1.08 |
| 10 | 0.42 | 1.08 |
| 13 | -0.64 | 1.08 |
| 20 | 0.11 | 1.08 |
| 21 | 0.21 | 1.08 |
| 22 | 0.21 | 1.08 |
| 26 | 0.42 | 1.08 |
| 27 | -0.09 | 1.08 |
| 29 | -0.09 | 1.08 |
| 30 | -0.64 | 1.08 |
If, for whatever reason, you want to reverse-identify specific individuals from the internal codes, you can match their codes:
## individuals of interest in internal codification
idx <- c(3, 5, 9)
## original codes
(match(idx, map.codes))## [1] 9 21 30