This repository is principally based on Schlegel et al. "Whole-brain annotation and multi-connectome cell typing of Drosophila", Nature (2024) which reports the systematic annotation of the FlyWire female adult fly brain (FAFB) connectome. The annotation data will be available here for download and have also been contributed to the https://codex.flywire.ai portal and FAFB-FlyWire CATMAID spaces.
The annotations collated here are used by the fafbseg-py Python and the fafbseg R package to enable programmatic analysis of the FlyWire dataset.
We're generally happy to consider contributions from the community to update/improve existing or add entirely new annotations. Please see the "How to contribute" section for details.
/supplemental_files/Supplemental_file1_neuron_annotations.tsvcontains neuron annotations for flow, superclass, cell class, nerve, lineage, side, morphology groups, neurotransmitter and VirtualFlyBrain IDs; this file is the basis for annotations infafbseg-py/supplemental_files/Supplemental_file2_non_neuron_annotations.tsvcontains annotations for non-neuronal objects such as trachae and glia/supplemental_files/Supplemental_file3_hemilineages_clustering.csvcontains details on the NBLAST clustering of hemilineages that generated the morphology groups/supplemental_files/Supplemental_file4_summary_with_ngl_links.csvcontains a summary for each hemilineage including neuroglancer links to view them/supplemental_files/Supplemental_file5_hemibrain_meta.csvcontains meta data for hemibrain (v1.2.1) pulled from neuPrint with some additional columns (e.g.side) used in our analyses
See here for detailed explanations for each column in these spreadsheets.
/code/annotation_counts.ipynbcontains examples of reading the annotation data and extracting numbers/counts used in the paper
To aid a number of analyses, hemibrain meshes were mapped into FlyWire (FAFB14.1) space. These can be co-visualised within neuroglancer for example by following this link: https://tinyurl.com/flywire783. This also enables direct querying of both our flywire annotations and hemibrain annotations from within neuroglancer to efficiently find and compare cell types.
High-resolution supplemental videos have been uploaded to YouTube:
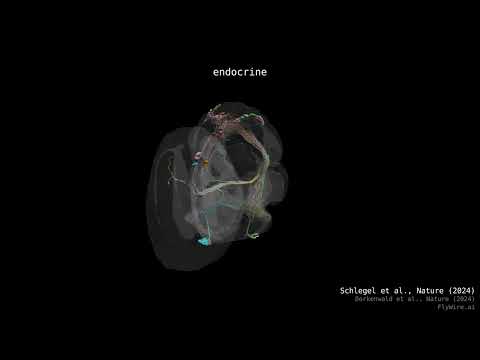
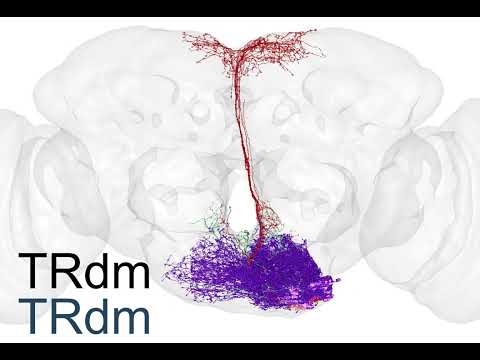
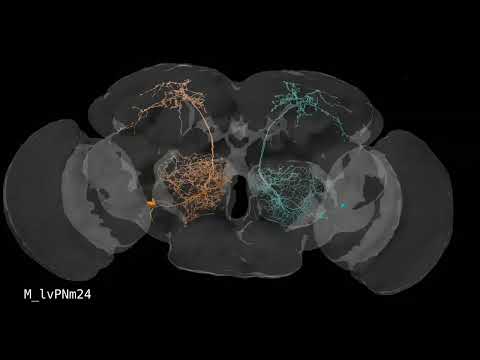
Supplemental Video 1: Rendering of all FlyWire neurons.
Supplemental Video 2: Rendering of neurons by superclass.
Supplemental Video 3: Hemilineage slides show.
Supplemental Video 4: FlyWire hemibrain cell type matches.
Skeletons and NBLAST scores are too large to be deposited on Github. Instead they are available for download via Zenodo: https://doi.org/10.5281/zenodo.10877326
Synapses table and edge list can be downloaded from https://zenodo.org/records/10676866 (provided by Sven Dorkenwald & FlyWire.ai).
All software used in this paper is open-source and available through Github. Some of it was specifically developed for working with FlyWire data. Please open an issue in the respective repository if you have questions or run into problems.
| Name | Description |
|---|---|
| navis | Analysis and visualisation of neurons. Used e.g. for NBLAST. |
| navis-flybrains | Used to transform data between template spaces (e.g. from hemibrain to FlyWire). |
| fafbseg-py | Query and analyse FlyWire data (segmentation, meshes, skeletons, annotations). |
| cocoa | Analysis suite for comparative connectomics. Enables e.g. hemibrain-FlyWire connectivity clustering. |
| neuprint-python | Query neuPrint instances (e.g. for the hemibrain). Developed by FlyEM (Janelia Research Campus). |
The recommended entry point for Python is fafbseg-py.
| Name | Description |
|---|---|
| natverse | Analysis suite with a focus on neuroanatomical data. |
| fafbseg | Query and analyse FlyWire data (segmentation, meshes, skeletons, annotations). |
| coconatfly | Analysis suite for Drosophila comparative connectomics. Enables hemibrain-FlyWire connectivity clustering. See also coconat. |
| neuprintr | Query neuPrint instances (e.g. for the hemibrain) |
The recommended entry point for R is coconatfly.
Skeletons, connectivity and annotations for FlyWire neurons have been imported into a CATMAID instance publicly available at https://fafb-flywire.catmaid.org/. This allows the interactive exploration and analysis of the data.
@article {Schlegel2024,
author = {Philipp Schlegel and Yijie Yin and Alexander Shakeel Bates and Sven Dorkenwald and Katharina Eichler and Paul Brooks and Daniel S Han and Marina Gkantia and Marcia dos Santos and Eva J Munnelly and Griffin Badalamente and Laia Serratosa Capdevila and Varun Aniruddha Sane and Alexandra M F Fragniere and Ladann Kiassat and Markus William Pleijzier and Imaan F M Tamimi and Christopher R Dunne and Irene Salgarella and Alexandre Javier and Siqi Fang and Eric Perlman and Tom Kazimiers and Sridhar R Jagannathan and Arie Matsliah and Amy R Sterling and Szi-chieh Yu and Claire E McKellar and FlyWire Consortium and Marta Costa and H. Sebastian Seung and Mala Murthy and Volker Hartenstein and Davi D Bock and Gregory S X E Jefferis},
title = {Whole-brain annotation and multi-connectome cell typing of Drosophila},
year = {2024},
doi = {10.1038/s41586-024-07686-5},
publisher = {Springer Nature},
journal = {Nature}
}Because annotations are still evolving we will occasionally update them. Thanks to Github's versioning you can always go back to the state at a given time (e.g. at initial publication) using the tags!
2.1.0: Still based on the783materialization but with substantial updates and revisions to cell types and other annotations. This is the version reported on in the Schlegel et al., Nature (2024) and Dorkenwald et al., Nature (2024).2.0.0: First major revision of annotations. With this release annotations are now based principally on the updated FlyWire segmentation version783. It includes new cell types and cell classes for the majority of neurons in both central brain and optic lobes.1.1.0: Second release for the updated Schlegel et al. (2023) bioRxiv version. Principally based on FlyWire segmentation version630.1.0.0: First release matching the Schlegel et al. bioRxiv paper. Principally based on FlyWire segmentation version630.
We welcome all kinds of contributions. For example:
- reports of incorrect annotations (e.g. cell types), broken links, etc.
- suggestions for new/updated cell type information (e.g. from more recent publications)
If you already know what needs doing, feel free to fork & PR right away. When in doubt please open an issue so we can discuss the best way to address the issue.