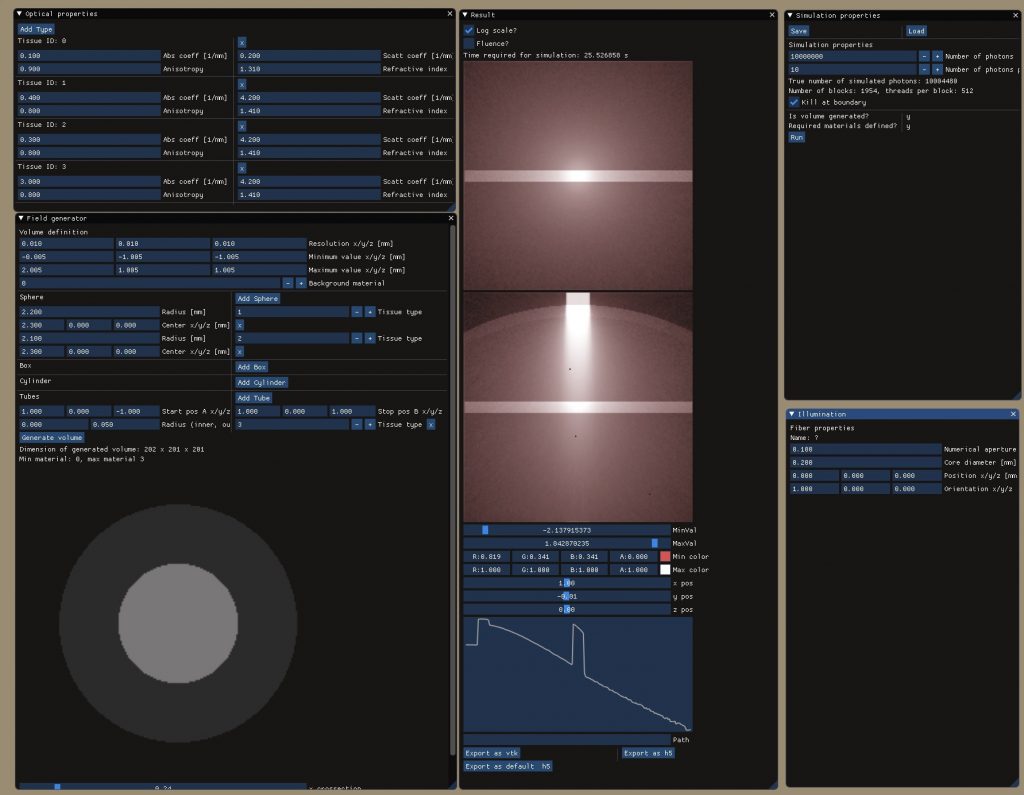
Disclaimer: This project is still in the development phase and while basic functionality was tested and validated, many features are still missing and require implementation (see Issue list).
Here we go, yet another toolbox for light simulations in highly scattering media like biological tissues. Completely unnecessary one might think. There are so many out there someone else might mention. Nevertheless, when I started to look for such a toolbox I was highly disappointed by the non-userfriendly versions which people published in the web over the last few years. Either using them is impossible due to a bad user interface or they perform incredibly slow because they are still running on a CPU.
This is my attempt to build a Monte Carlo simulation toolbox featuring CUDA acceleration on NVIDIA cards, a friendly user interface based on the amazing ImGui project, handling multilayered tissues with elegance, geometry creation on the fly and directly from the GUI, combining multiple illuminations profiles in a single simulation, and export to datatypes readable from different software like Python, MATLAB, etc. through the established h5 standard. The simulations are performed voxel by voxel relying on Code extensively tested on the CPU.
All settings are stored through the json file format which allows defining them through different interfaces and loading them into the GUI in a single click. Export of simulation datasets is available through h5 or to vtk for alterantive displaying or postprocessing.
Documentation is lacking for now. Lazy programmers often say that an intuitive user interface does not require too much documentation.
The code was written for and tested with Linux. Please send me a request if you do not know how to install it on your system and I will provide you with instructions.
Libraries / pacakges required:
hdf5general purpose library and file format for storing scientific datacudaandnvidiadriver (eventually you want to usenvidia-ltsif you uselinux-lts)- stuff required for imgui
cmakeandmakeglfw-x11,glewused to display stuffnlohmann-jsonsave settings of simulation to json file
Archlinux installation command
pacman -S nlohmann-json cuda nvidia hdf5 cmake make glfw-x11 glew
Ubuntu installation command
apt-get install libhdf5-dev nvidia-cuda-toolkit cmake make libglfw3-dev libglfw3-dev libglew-dev nlohmann-json3-dev libsdl2-dev git g++
On Ubuntu there is still a problem with the hdf5 library. I am working on it.
This software is written for Linux and requires CUDA. Switch on the terminal, cd to your favorite installation directory and run the following command cascade:
git clone git@github.com:hofmannu/yamct.git
cd yamct
git submodule init
git submodule update
mkdir Debug
cd Debug
cmake .. && make all && ./main_exp
Building in debugging mode
cmake -DCMAKE_BUILD_TYPE=Debug .. && make all && gdb main_exp
Make sure you have gdb installed.
I am actively working on this project. If you want any feature implemented (for example different geometrical shapes, export types, or illumination types) feel free to open an issue and I will get back to you as soon as possible.
Things which are on my ToDo list include
- Different illumination types (e.g. gaussian beam)
- Predefined tissue types to automatically load optical properties depending on used wavlenght
- Export to different file types including
mat - Testing for Windows and Ubuntu
- Documentation for settings defined in
jsonfile
On the website of OMLC you can find an extensive list of Literature about light simulation through Monte Carlo simulations as well as optical properties of different tissue types.