forked from PaddlePaddle/PaddleScience
-
Notifications
You must be signed in to change notification settings - Fork 0
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
【Hackathon 6th No.39】XPINN 迁移至 PaddleScience (PaddlePaddle#849)
* add XPINNs example * comment * add conf file * refine code * fix comment * fix data type * fix data type * Update examples/xpinn/plotting.py * Update examples/xpinn/conf/xpinn.yaml * refine code * refine doc * refine doc * fix doc * fix bugs --------- Co-authored-by: HydrogenSulfate <490868991@qq.com>
- Loading branch information
1 parent
452ba2f
commit ce050a3
Showing
5 changed files
with
1,151 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,184 @@ | ||
| # Extended Physics-Informed Neural Networks (XPINNs) | ||
|
|
||
| === "模型训练命令" | ||
|
|
||
| ``` sh | ||
| # linux | ||
| wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/XPINN/XPINN_2D_PoissonEqn.mat -P ./data/ | ||
|
|
||
| # windows | ||
| # curl https://paddle-org.bj.bcebos.com/paddlescience/datasets/XPINN/XPINN_2D_PoissonEqn.mat --output ./data/XPINN_2D_PoissonEqn.mat | ||
|
|
||
| python xpinn.py | ||
|
|
||
| ``` | ||
|
|
||
| ## 1. 背景简介 | ||
|
|
||
| 求解偏微分方程(PDE)是一类基础的物理问题,随着人工智能技术的高速发展,利用深度学习求解偏微分方程成为新的研究趋势。[XPINNs(Extended Physics-Informed Neural Networks)](https://doi.org/10.4208/cicp.OA-2020-0164)是一种适用于物理信息神经网络(PINNs)的广义时空域分解方法,以求解任意复杂几何域上的非线性偏微分方程。 | ||
|
|
||
| XPINNs 通过广义时空区域分解,有效地提高了模型的并行能力,并且支持高度不规则的、凸/非凸的时空域分解,界面条件是简单的。XPINNs 可扩展到任意类型的偏微分方程,而不论方程是何种物理性质。 | ||
|
|
||
| 精确求解高维复杂的方程已经成为科学计算的最大挑战之一,XPINNs 的优点使其成为模拟复杂方程的适用方法。 | ||
|
|
||
| ## 2. 问题定义 | ||
|
|
||
| 二维泊松方程: | ||
|
|
||
| $$ \Delta u = f(x, y), x,y \in \Omega \subset R^2$$ | ||
|
|
||
| ## 3. 问题求解 | ||
|
|
||
| 接下来开始讲解如何将问题一步一步地转化为 PaddleScience 代码,用深度学习的方法求解该问题。 | ||
| 为了快速理解 PaddleScience,接下来仅对模型构建、方程构建、计算域构建等关键步骤进行阐述,而其余细节请参考 [API文档](../api/arch.md)。 | ||
|
|
||
| ### 3.1 数据集下载 | ||
|
|
||
| 如下图所示,数据集包含计算域的三个子区域的数据:红色区域的边界和残差点;黄色区域的界面;以及绿色区域的界面。 | ||
|
|
||
| <figure markdown> | ||
| 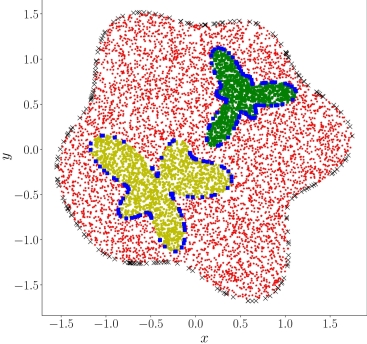 | ||
| <figcaption>二维泊松方程的三个子区域</figcaption> | ||
| </figure> | ||
|
|
||
| 计算域的边界表达式如下。 | ||
|
|
||
| $$ \gamma =1.5+0.14 sin(4θ)+0.12 cos(6θ)+0.09 cos(5θ), θ \in [0,2π) $$ | ||
|
|
||
| 红色区域和黄色区域的界面的表达式如下。 | ||
|
|
||
| $$ \gamma_1 =0.5+0.18 sin(3θ)+0.08 cos(2θ)+0.2 cos(5θ), θ \in [0,2π)$$ | ||
|
|
||
| $$ \gamma_2 =0.34+0.04 sin(5θ)+0.18 cos(3θ)+0.1 cos(6θ), θ \in [0,2π) $$ | ||
|
|
||
| 执行以下命令,下载并解压数据集。 | ||
|
|
||
| ``` sh | ||
| wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/XPINN/XPINN_2D_PoissonEqn.mat -P ./data/ | ||
| ``` | ||
|
|
||
| ### 3.3 模型构建 | ||
|
|
||
| 在本问题中,我们使用神经网络 `MLP` 作为模型,在模型代码中定义三个 `MLP` ,分别作为三个子区域的模型。 | ||
|
|
||
| ``` py linenums="301" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:301:302 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| 模型训练时,我们将使用 XPINN 方法分别计算每个子区域的模型损失。 | ||
|
|
||
| <figure markdown> | ||
| 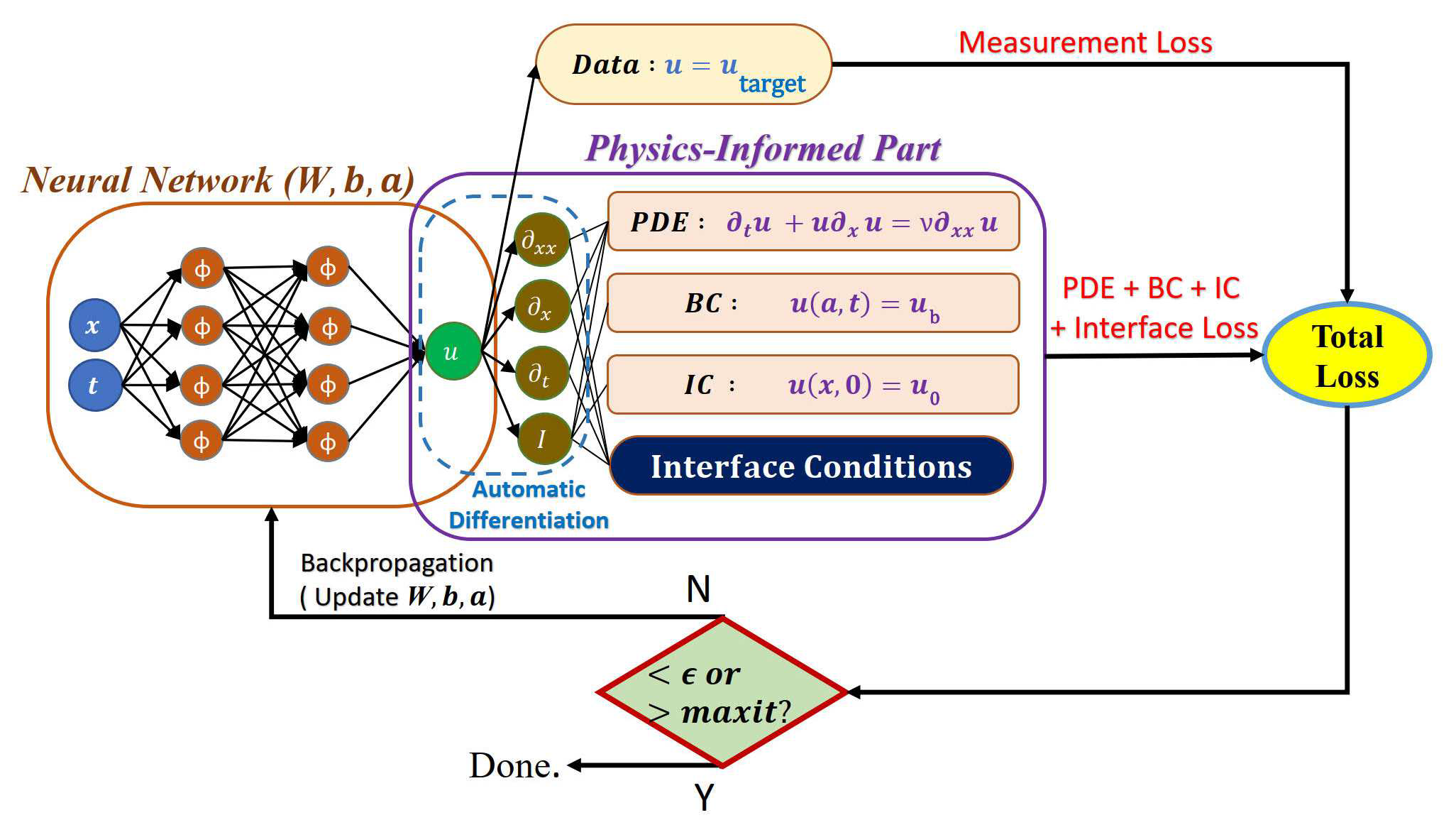 | ||
| <figcaption>XPINN子网络的训练过程</figcaption> | ||
| </figure> | ||
|
|
||
| ### 3.4 约束构建 | ||
|
|
||
| 在本案例中,我们使用监督数据集对模型进行训练,因此需要构建监督约束。 | ||
|
|
||
| 在定义约束之前,我们需要指定数据集的路径等相关配置,将这些信息存放到对应的 YAML 文件中,如下所示。 | ||
|
|
||
| ``` yaml linenums="43" | ||
| --8<-- | ||
| examples/xpinn/conf/xpinn.yaml:43:44 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| 接着定义训练损失函数的计算过程,调用 XPINN 方法计算损失,如下所示。 | ||
|
|
||
| ``` py linenums="130" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:130:191 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| 最后构建监督约束,如下所示。 | ||
|
|
||
| ``` py linenums="304" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:304:311 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| ### 3.5 超参数设定 | ||
|
|
||
| 设置训练轮数等参数,如下所示。 | ||
|
|
||
| ``` yaml linenums="83" | ||
| --8<-- | ||
| examples/xpinn/conf/xpinn.yaml:83:88 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| ### 3.6 优化器构建 | ||
|
|
||
| 训练过程会调用优化器来更新模型参数,此处选择较为常用的 `Adam` 优化器。 | ||
|
|
||
| ``` py linenums="337" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:337:338 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| ### 3.7 评估器构建 | ||
|
|
||
| 在训练过程中通常会按一定轮数间隔,用验证集(测试集)评估当前模型的训练情况,因此使用 `ppsci.validate.SupervisedValidator` 构建评估器。 | ||
|
|
||
| ``` py linenums="324" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:324:335 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| 评估指标为预测结果和真实结果的 RMSE 值,这里需自定义指标计算函数,如下所示。 | ||
|
|
||
| ``` py linenums="194" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:194:219 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| ### 3.8 模型训练 | ||
|
|
||
| 完成上述设置之后,只需要将上述实例化的对象按顺序传递给 `ppsci.solver.Solver`,然后启动训练。 | ||
|
|
||
| ``` py linenums="340" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:340:357 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| ### 3.9 结果可视化 | ||
|
|
||
| 训练完毕之后程序会对测试集中的数据进行预测,并以图片的形式对结果进行可视化,如下所示。 | ||
|
|
||
| ``` py linenums="360" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py:360:384 | ||
| --8<-- | ||
| ``` | ||
|
|
||
| ## 4. 完整代码 | ||
|
|
||
| ``` py linenums="1" title="cfdgcn.py" | ||
| --8<-- | ||
| examples/xpinn/xpinn.py | ||
| --8<-- | ||
| ``` | ||
|
|
||
| ## 5. 结果展示 | ||
|
|
||
| 下方展示了对计算域中每个点的预测值结果、参考结果和相对误差。 | ||
|
|
||
| <figure markdown> | ||
| 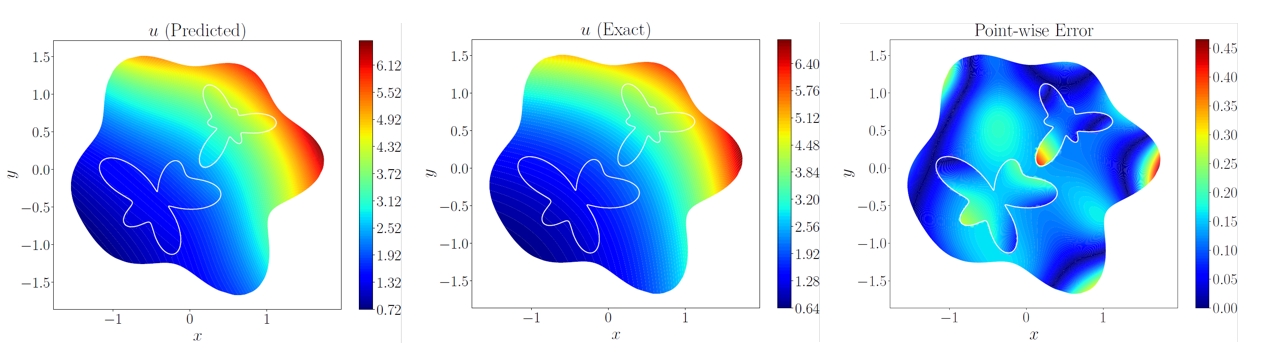 | ||
| <figcaption>预测结果和参考结果的对比</figcaption> | ||
| </figure> | ||
|
|
||
| 可以看到模型预测结果与真实结果相近,若增大训练轮数,模型精度会进一步提高。 | ||
|
|
||
| ## 6. 参考文献 | ||
|
|
||
| * [A.D.Jagtap, G.E.Karniadakis, Extended Physics-Informed Neural Networks (XPINNs): A Generalized Space-Time Domain Decomposition Based Deep Learning Framework for Nonlinear Partial Differential Equations, Commun. Comput. Phys., Vol.28, No.5, 2002-2041, 2020.](https://doi.org/10.4208/cicp.OA-2020-0164) |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,122 @@ | ||
| hydra: | ||
| run: | ||
| # dynamic output directory according to running time and override name | ||
| dir: outputs_xpinn/${now:%Y-%m-%d}/${now:%H-%M-%S}/${hydra.job.override_dirname} | ||
| job: | ||
| name: ${mode} # name of logfile | ||
| chdir: false # keep current working direcotry unchaned | ||
| config: | ||
| override_dirname: | ||
| exclude_keys: | ||
| - TRAIN.checkpoint_path | ||
| - TRAIN.pretrained_model_path | ||
| - EVAL.pretrained_model_path | ||
| - mode | ||
| - output_dir | ||
| - log_freq | ||
| callbacks: | ||
| init_callback: | ||
| _target_: ppsci.utils.callbacks.InitCallback | ||
| sweep: | ||
| # output directory for multirun | ||
| dir: ${hydra.run.dir} | ||
| subdir: ./ | ||
|
|
||
| # general settings | ||
| mode: train # running mode: train/eval | ||
| seed: 134 | ||
| output_dir: ${hydra:run.dir} | ||
| log_freq: 20 | ||
|
|
||
| # model settings | ||
| MODEL: | ||
| num_boundary_points: 200 # Boundary points from subdomain 1 | ||
| num_residual1_points: 5000 # Residual points in three subdomain 1 | ||
| num_residual2_points: 1800 # Residual points in three subdomain 2 | ||
| num_residual3_points: 1200 # Residual points in three subdomain 3 | ||
| num_interface1: 100 # Interface points along the two interfaces | ||
| num_interface2: 100 | ||
| layers1: [2, 30, 30, 1] | ||
| layers2: [2, 20, 20, 20, 20, 1] | ||
| layers3: [2, 25, 25, 25, 1] | ||
|
|
||
| # set training data file | ||
| DATA_FILE: "./data/XPINN_2D_PoissonEqn.mat" | ||
|
|
||
| # training settings | ||
| TRAIN: | ||
| input_keys: | ||
| [ | ||
| "residual1_x", | ||
| "residual1_y", | ||
| "residual2_x", | ||
| "residual2_y", | ||
| "residual3_x", | ||
| "residual3_y", | ||
| "interface1_x", | ||
| "interface1_y", | ||
| "interface2_x", | ||
| "interface2_y", | ||
| "boundary_x", | ||
| "boundary_y", | ||
| ] | ||
| label_keys: ["boundary_u_exact"] | ||
| alias_dict: | ||
| { | ||
| "residual1_x": "x_f1", | ||
| "residual1_y": "y_f1", | ||
| "residual2_x": "x_f2", | ||
| "residual2_y": "y_f2", | ||
| "residual3_x": "x_f3", | ||
| "residual3_y": "y_f3", | ||
| "interface1_x": "xi1", | ||
| "interface1_y": "yi1", | ||
| "interface2_x": "xi2", | ||
| "interface2_y": "yi2", | ||
| "boundary_x": "xb", | ||
| "boundary_y": "yb", | ||
| "boundary_u_exact": "ub", | ||
| "residual_u_exact": "u_exact", | ||
| "residual2_u_exact": "u_exact2", | ||
| "residual3_u_exact": "u_exact3", | ||
| } | ||
| epochs: 501 | ||
| iters_per_epoch: 1 | ||
| save_freq: 50 | ||
| eval_during_train: true | ||
| eval_freq: 50 | ||
| learning_rate: 0.0008 | ||
| pretrained_model_path: null | ||
| checkpoint_path: null | ||
|
|
||
| # evaluation settings | ||
| EVAL: | ||
| label_keys: | ||
| [ | ||
| "boundary_u_exact", | ||
| "residual_u_exact", | ||
| "residual2_u_exact", | ||
| "residual3_u_exact", | ||
| ] | ||
| alias_dict: | ||
| { | ||
| "residual1_x": "x_f1", | ||
| "residual1_y": "y_f1", | ||
| "residual2_x": "x_f2", | ||
| "residual2_y": "y_f2", | ||
| "residual3_x": "x_f3", | ||
| "residual3_y": "y_f3", | ||
| "interface1_x": "xi1", | ||
| "interface1_y": "yi1", | ||
| "interface2_x": "xi2", | ||
| "interface2_y": "yi2", | ||
| "boundary_x": "xb", | ||
| "boundary_y": "yb", | ||
| "boundary_u_exact": "ub", | ||
| "residual_u_exact": "u_exact", | ||
| "residual2_u_exact": "u_exact2", | ||
| "residual3_u_exact": "u_exact3", | ||
| } | ||
| batch_size: 1 | ||
| pretrained_model_path: null | ||
| eval_with_no_grad: false |
Oops, something went wrong.