A Python package to interact with fasting logs from apps like Zero.
- Free software: MIT license
- Documentation: https://jbpauly.github.io/fasting
Fasting is a technique used to improve metabolic health, reduce inflammation, rebalance gut health, lose weight, increase longevity, improve mental clarity and focus. There's various forms of fasting, with Intermittent Fasting (IF) likely being the most common. You can read about IF and other types of fasting on the Zero blog. During extended periods of fasting, our bodies "flip a metabolic switch" from burning glucose to burning fatty acids and ketones.
The metabolic switch typically takes place in a later phase of fasting, when the glycogen stores contained in the liver are depleted and adipose tissue (fat) begins to increase fatty acids and glycerol. While the switch generally occurs between 12 and 36 hours after stopping eating, it can depend upon the total liver glycogen content when the person started the fast, as well as the person’s energy expenditures during the fast.
Josh Clemente, a Levels co-founder, has shared the benefits of IF on his metabolic fitness on the Levels blog.
During periods of extended fasts, Josh's average glucose gently trends down into the optimal range and remains there with only 2-3% variation for days at a time.
This package is part of the Digital Biomarker Discovery Pipeline (DBDP).
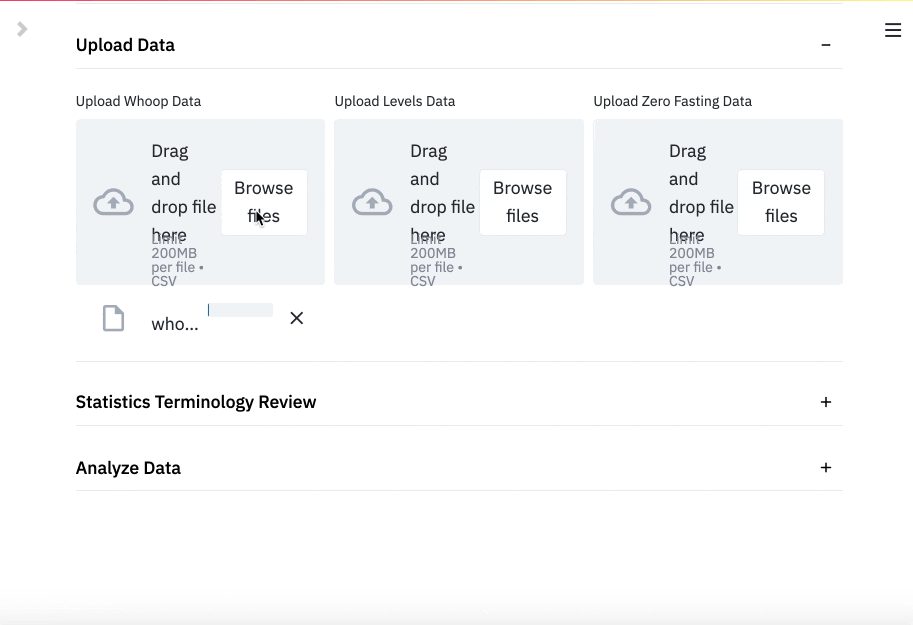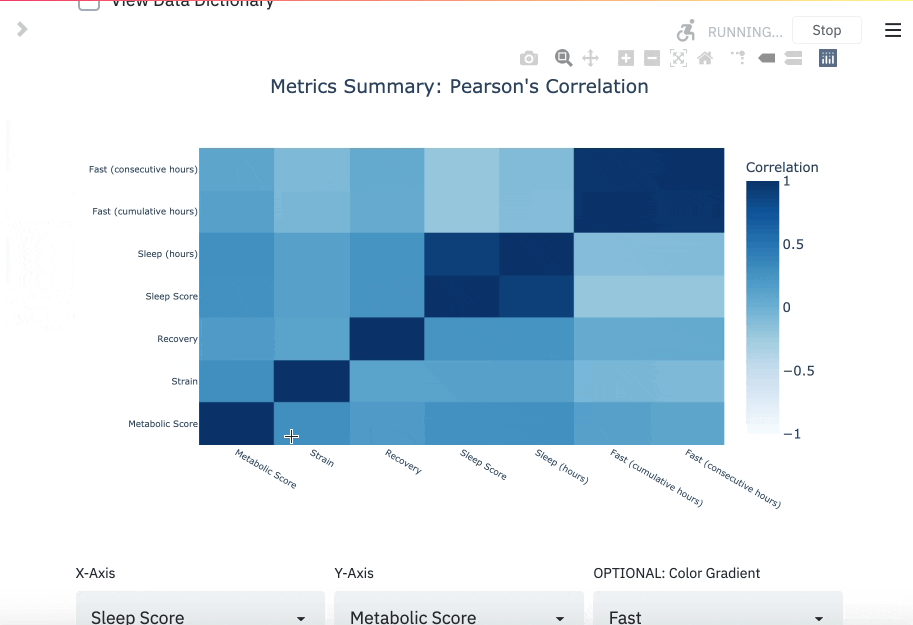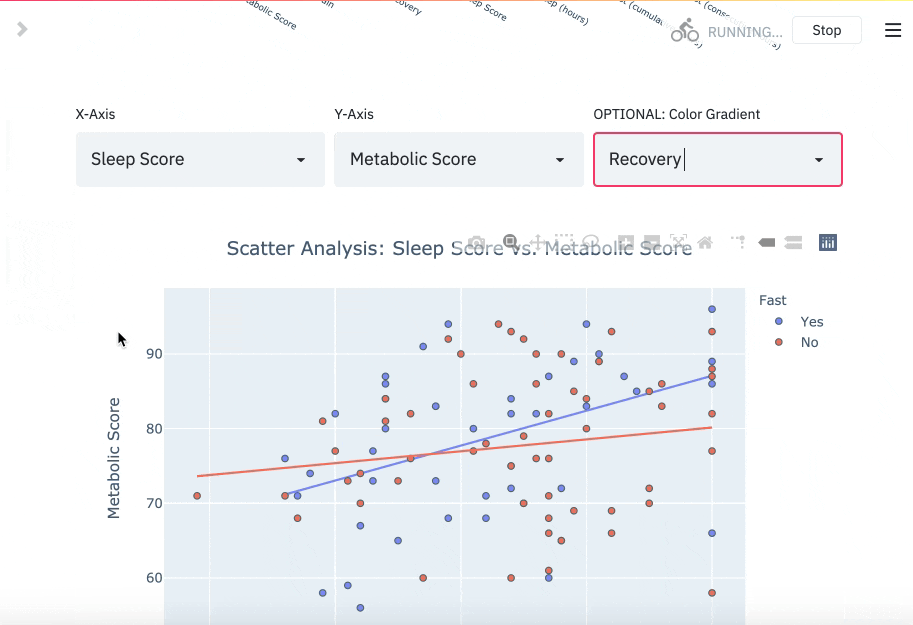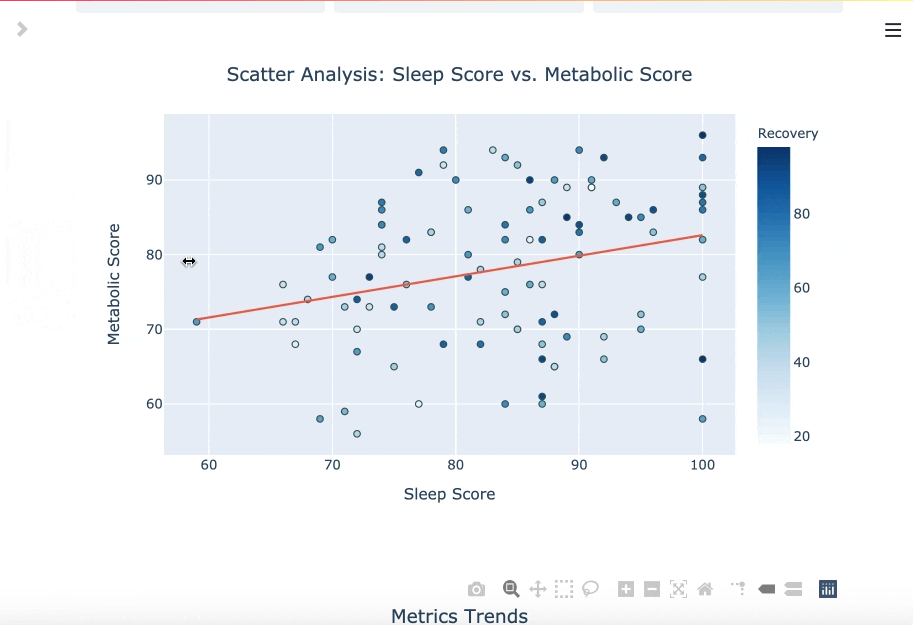
You can read more about the DBDP here. If you track your fasts, there's a chance you also track biomarkers with wearables and other hardware in the modern health stack. The goal of this package is to enable you to utilize your data for your own purpose, like cross analyzing fasting performance with other biomarkers.
Functionality around fasting logs was originally built for the Metabolic Health Analysis app.
Part of the app's software was pulled out and enhanced to create this stand-alone FASTING package. Checkout the app repo and the DBDP for ideas and use cases around your own biomarker analysis.
This section provides a quick preview of some of the package features. Check out the full API documentation and getting started tutorial for more information.
This package is supported by Python 3.7+.
pip install fasting
Export format from Zero:
| Date | Start | End | Hours | Night Eating |
|---|---|---|---|---|
| 1/17/21 | 19:15 | 12:00 | 16 | 1 |
| 1/16/21 | 19:45 | 10:14 | 14 | 2 |
| 1/15/21 | 21:00 | 10:00 | 13 | 3 |
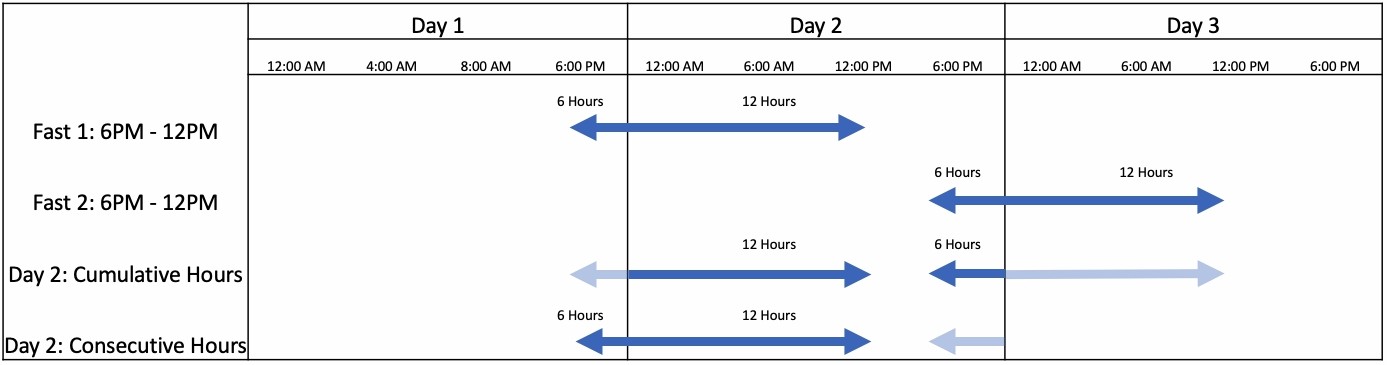
Discrete Logs:
discrete_logs = fasting.quantify.zero_fasts(zero_export.csv)
| fast | start_dt | end_dt |
|---|---|---|
| 0 | 2021-01-14 19:00:00 | 2021-01-15 11:22:00 |
| 1 | 2021-01-15 21:00:00 | 2021-01-16 10:00:00 |
| 2 | 2021-01-16 19:45:00 | 2021-01-17 10:14:00 |
Continuous Log:
continuous_log = fasting.quantify.continuous_fasts(discrete_logs)
| datetime | fasting status |
|---|---|
| 2021-01-14 19:00:00 | 1 |
| 2021-01-14 19:01:00 | 1 |
| 2021-01-14 19:02:00 | 1 |
cumulative_hours = quantify.daily_cumulative_hours(continuous_log)
max_consecutive_hours = quantify.daily_max_consecutive_hours(continuous_log)
Impact of time restricted feeding (TRF) on metabolic health
- Two groups
- Group 1: 6 hour feeding window (8am-2pm), TRF group
- Group 2: 12 hour feeding window (8am-8pm), control group
- Overweight, non-diabetic individuals
Result / Findings:
- Engaging in time restricted feeding for just 4 days can lower fasting glucose, fasting insulin, and mean glucose levels significantly
- TRF group demonstrated lower post meal glucose spikes, despite eating the same meals
Relationship between time of day of eating and metabolic health
Result / Findings: Eating food later in the evening will cause significant increase in insulin and glucose levels compared to eating the same meal in the morning
Impact of IF on controlling type 2 diabetes
- 3 participants with type 2 diabetes
- 24 hour fasts 3-4 days per week
Result / Findings:
- All participants meaningfully reversed diabetes in as little as 7 months
- One participant regained enough insulin sensitivity to get off high doses of insulin medication within 3 weeks
This package was created with Cookiecutter and the giswqs/pypackage project template.