This is an OpenSource project collaborating with a healthcare mutual insurance company (data provider). The aim of this project is to build a pipeline to classify knee frontal radiography images according to Schatzker classification (from 0 to 6) from DICOM files. However, as first approach it used a binary classification of fractured knee (Schatzker classification different than 0).
Medical imaging projects as this one have several features that should be taken into account:
-
Lack of datasets/samples for specific task
-
Annotation requires expertise on traumatology field, hence it is limited
-
Data is highly imbalanced towards non-fractured knees
-
Medical images require special treatments
In order to tackle these issues we implemented the following methods on PyTorch using Fastai platform:
-
Algorithm based:
- Transfer Learning on several steps (first ImageNet and then using DICOM metadata for pretraining model)
- Self-Supervised Learning (
pytorch-lightningpackage) for state-of-the-art transfer learning - Semi-Supervised Learning (MixMatch and FixMatch own implementations) to be able to train model without full dataset annotated
- Organize experiments for Hyperparameters search (MLFlow or Ax-BoTorch)
-
Data based:
- DataLoader sampler which allows to have a minimum of samples of all the labels in each batch
- Use of specific losses to be robust to imbalance datasets
- Active Learning to select most interesting files to annotate
-
Image processing:
- Preprocesses to transform DICOM files into images independently of its metadata
- Adaptative equalization of histogram to normalize bimodal data
- Improve Knee localization to center-crop images on relevant part
- Clean background to avoid overfitting on noise
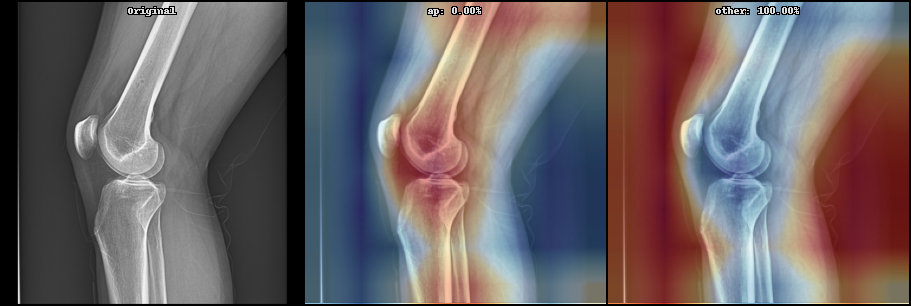
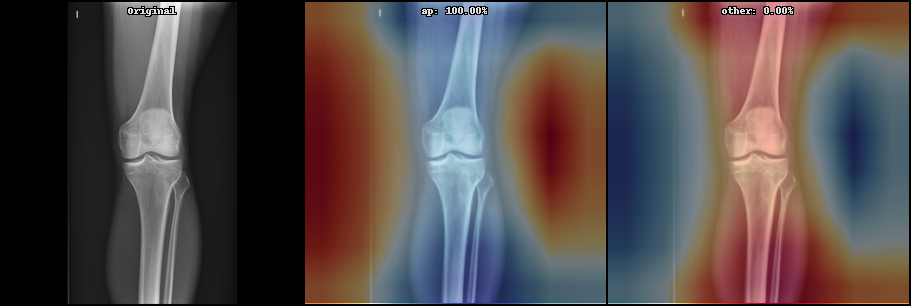
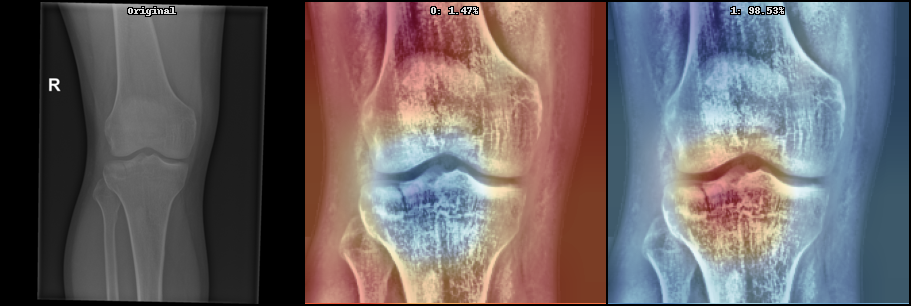
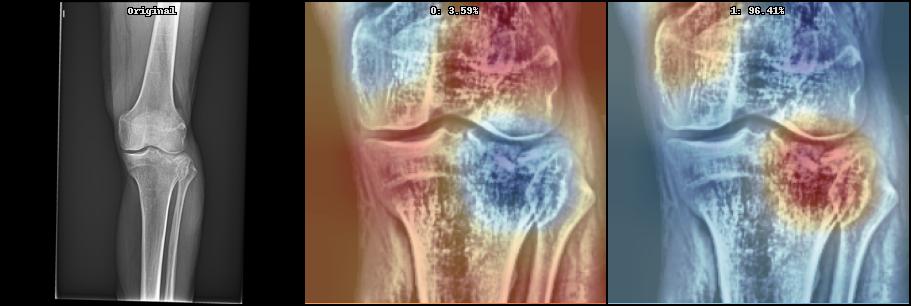
- Model interpretation with GradCAM or Layer Relevance Propagation
- Use several channels for different views of the radiography
See below some examples these image processing:
For more information regarding the implemented preprocesses please see Preprocessing README or the related API documentation here
Below can be seen some initial results on the current state (May 2021) on supervised learning:
All the notebooks are prepared to be directly used on Google Colab, then if you are going to be using Google Colab you do not have to worry at all.
In case you would like to reuse code for setting up your notebook please look on colab_pip_setup.ipynb and/or colab_conda_setup.ipynb. This last one is for the automatic conda installations on Google Colab. The gdcm package which is required to read all kind of DICOM files can only be installed with conda. Hence 0_eda.ipynb and 3_preprocess.ipynb notebooks which read DICOM files requires of this kind of installation.
Add conda-forge channel:
conda config --add channels conda-forge
Create environment:
conda create -n radiology_ai python=3.6 --yes -q
conda activate radiology_ai
Install environment:
conda install -n radiology_ai --override-channels -c main -c conda-forge -c fastai -c pytorch pytorch=1.6.0 fastai fastcore=1.0.9 pydicom gdcm=2.8.4 kornia=0.3.0 scikit-image scikit-learn pandas ipykernel ipywidgets tensorboard openpyxl black --yes -q
Install EfficientNet for Pytorch and Pytorch Ligthning Bolts:
pip install efficientnet_pytorch opencv-python
Install Pytorch Lignthning Bolts (due to an issue not yet solved on release better to use this than pip install pytorch-lightning):
pip install git+https://github.com/PytorchLightning/pytorch-lightning-bolts.git@master --upgrade
pip install https://github.com/lluissalord/pytorch-lightning-bolts/archive/issue_436_simCLR_IndexError.zip --upgrade
Install fastai2_extensions (renamed as amalgam) to use gradCAM visualizations:
pip install https://github.com/lluissalord/fastai2_extensions/archive/fix_to_cuda.zip palettable
API Documentation on lluissalord.github.io/radiology_ai/
-
AttributeError: '_FakeLoader' object has no attribute 'persistent_workers':
pip install --upgrade fastai -
Progress bar issues using VSCode:
conda install "ipython>=6.0.0"