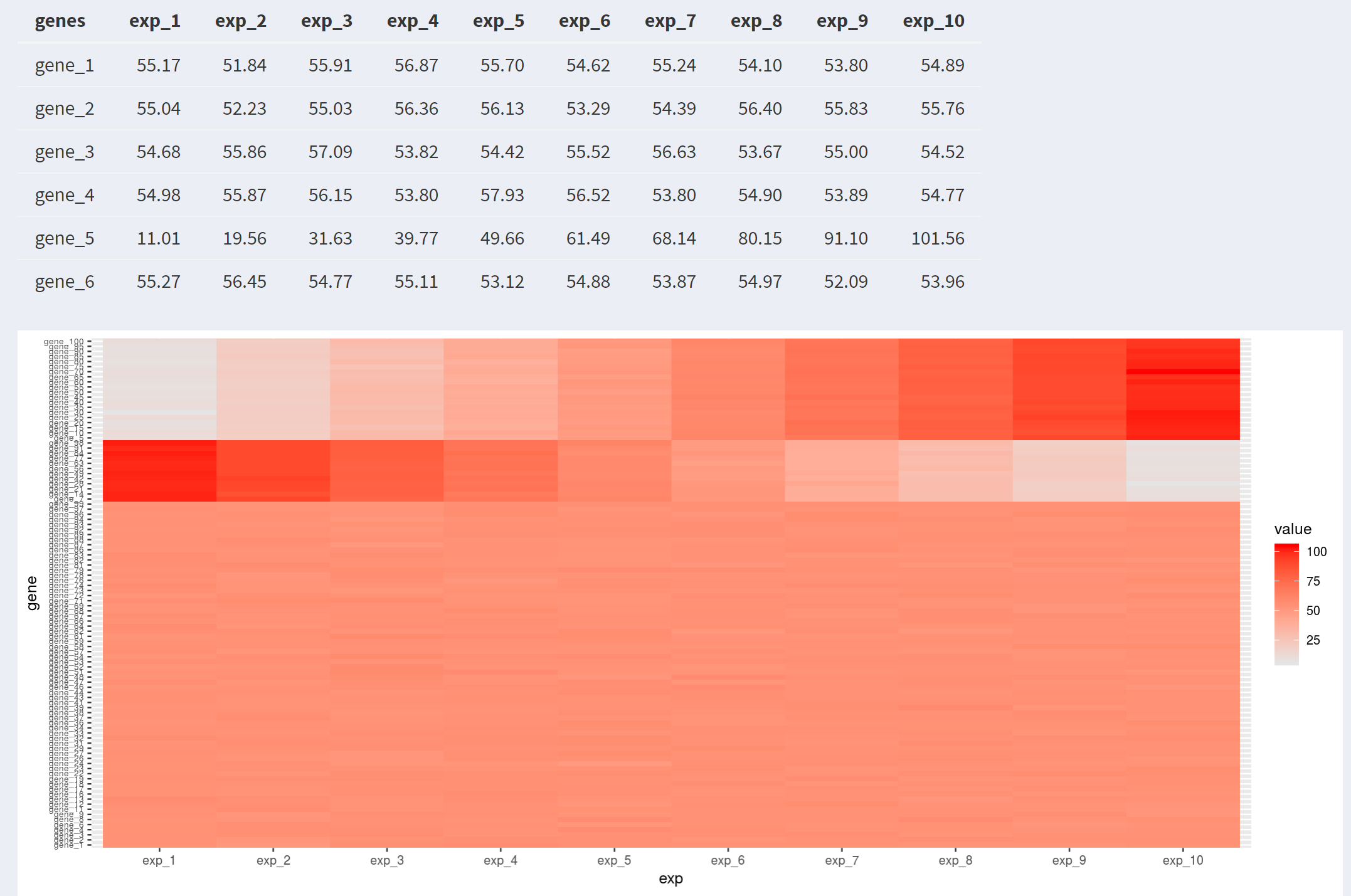
This file contains pre-normalized expression values for 100 genes over 10 time points. Most genes have a stable background expression level, but some special genes show increased expression over the timecourse and some show decreased expression.
This shiny app https://luchao-qi.shinyapps.io/shiny_clustering/ can
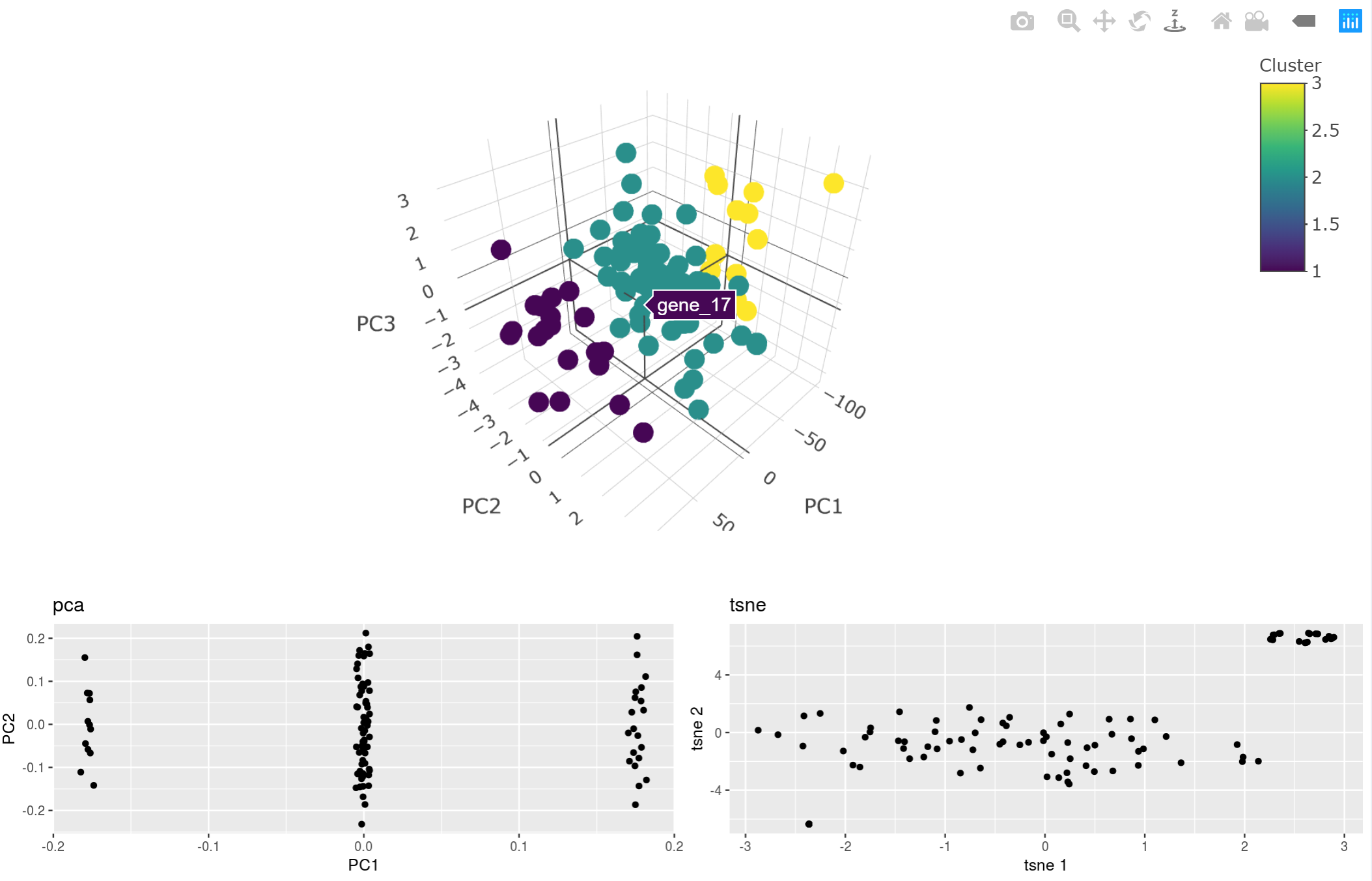
- Cluster the genes using K-means clustering
- Create a heatmap of the expression matrix. Order the genes by cluster, but keep the time points in numerical order.
- Compare the performance among PCA, T-sne, Umap analysis
- Import a txt file and format it to data frame.
- Choose the number of clusters you want.
- Visualize different clustering results so you can compare different machine learning algorithms!
- Upload your own data or download sample data on the left sidebar
- Enter the number of clusters
- Results
Free Software, Hell Yeah!