This repository contains code for analyzing how word usage differs across related groups of text data. The corresponding publication is
Also, check out the NIPS 2017 Spotlight Video:
The code in this repository contains 3 models you can fit to grouped text data:
- global Bernoulli embeddings (no variations between groups)
- hierarchical Bernoulli embeddings (embeddings share statistical strength through hierarchical prior)
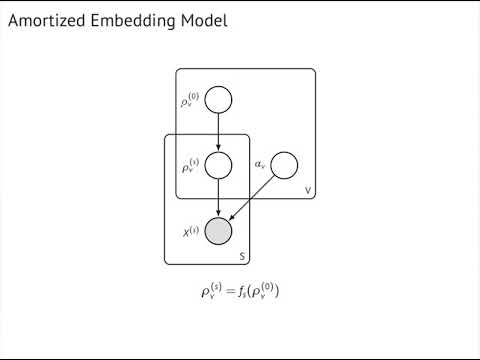
- amortized Bernoulli embeddings (embeddings share statistical strength through amortization)
We use these models to analyze how the language of Senators differs according to their home state and party affiliation and how scientific language varies in differerent sections of the ArXiv.
All code in this repo has been run and tested on Linux, with Python 2.7 and Tensorflow 1.3.0.
The input format, and how to prepare the data so it has the required input format is described in the dat/ subfolder of this repo. Follow the instructions in the README, code is provided.
To fit the models, go into the source folder (src/) and run
python main.py --fpath [path/to/data]
substitute the path to the folder where you put the data for [path/to/data].
For example, after running the python scripts step_1 through step_4 in the dat folder, you can run python main.py --fpath lorem_ipsum --K 5 to run Bernoulli embeddings on the provided test data dat/lorem_ipsum.
For all commandline options run:
python main.py --help
For fastest convergence we recommend a 2-step training procedure. Step 1 fits a Bernoulli embedding which is then used to initialize a structured embedding.
First run
python main.py --fpath [path/to/data]
This executes Bernoulli embeddings without structure. The script uses the current timestamp to create a folder where the results are saved ([path/to/results/]). We will use these results to initialize the structured embeddings:
python main.py --hierarchical True --fpath [path/to/data] --init [path/to/result]/variational0.dat
or
python main.py --amortized True --fpath [path/to/data] --init [path/to/result]/variational0.dat
Make sure to use the same --K for both runs.