-
Notifications
You must be signed in to change notification settings - Fork 3.8k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[dask] Distributed training sometimes produces very high leaf values #4026
Comments
|
FWIW this doesn't happen with dask-lightgbm. I'll try to find where the problem is. |
|
This actually seems to be related to lightgbm itself. I just pasted the original dask-lightbm code in |
|
How much data (rows) you used? You can try to construct a dataset on a single node first, via Dataset.construct(), then save it to binary format, Dataset.save_binary(). Then use the binary file for the distributed learning. |
|
The example I posted generates 20 time series between 300 and 700 rows each, so on average they would be around 10,000 rows. However the issue persists when having more data, I just changed the 20 to 1,000 and trained on 500,000 rows and reproduced the issue. I'm using two machines, I also noticed that when adding more machines the values on the nodes go even higher (even with big datasets). I'm trying to track down exactly where this originated because building from 2.3.1 (8364fc3) works fine, however with some of the later changes, (da91c61, d0bec9e) I get this error: |
|
@jmoralez I think it is better to fix (use the same) dataset first, constructing it by all rows. |
|
Do you mean using the CLI? |
These are all python interfaces. |
|
I've constructed the dataset and saved it to binary format. I'm just confused about
|
|
After you save binary file, you can copy it to the nodes for distributed learning. This will force the distributed learning to use the same dataset as the single-machine mode. |
|
Thank you. I just trained using the CLI on my local machine with two "machines" using the binary file and then loaded the booster in python to see the leaf values and they seem normal. Do you have any suggestions for the dask case? |
|
okay, so the problem indeed happens in Dataset construct. To ultimately fix this problem, we should add an "accurate" mode, for feature bucketing. This will significantly slow-down the pre-processing stage before training, due to heavy communications. also cc @shiyu1994 to implement "accurate" mode The algorithm of "accurate" mode is like:
|
|
I should point out that this doesn't happen with all datasets. For example, using |
|
@jmoralez |
|
I get high values as well |
|
I think it has to do with low variance features. In this example I create for example |
|
@jmoralez did you shuffle the data by rows? |
|
I did try it but it doesn't help |
|
I just tried using |
|
cc @shiyu1994 for possible bugs. |
|
@jameslamb I'm debugging this. Could you tell me how to make dask regressor print the Warning messages from the C++ code? I seems that setting neither |
|
@shiyu1994 sure, I think I can help. This answer uses a modified version of the code from https://github.com/microsoft/LightGBM/blob/master/examples/python-guide/dask/regression.py (just added
python
import dask.array as da
from distributed import Client, LocalCluster
from sklearn.datasets import make_regression
import lightgbm as lgb
cluster = LocalCluster(n_workers=2)
client = Client(cluster)
X, y = make_regression(n_samples=1000, n_features=50)
dX = da.from_array(X, chunks=(100, 50))
dy = da.from_array(y, chunks=(100,))
dask_model = lgb.DaskLGBMRegressor(n_estimators=10, silent=False, verbose=2)
dask_model.fit(dX, dy)When I did this, I saw the following printed in my Python console after calling Are there other specific log messages that you think are missing? One other thing you can tryYou might be able to find more diagnostic information on the Dask dashboard, especially in the "Info" tab that can take you to additional worker logs. In that same Python REPL, you could get the address for the dashboard by running print(client.dashboard_link)Paste that into a browser and you'll be able to see the Dask diagnostic dashboard. But when I did that, I only saw Dask logs and nothing else from LightGBM. |
|
ok now I'm weirded out by the changing |
|
@jameslamb Thanks! I do find a bug in data parallel training. When values of the same feature in different machines differ a lot, or even have no overlap, a global optimal split can result in leaves with Even under such case, we are expected to provide a stable (perhaps not good enough) result. But the problem is, when a leaf has data count LightGBM/include/LightGBM/dataset.h Lines 470 to 478 in 6ad3e6e
This will not cause any problem in a single machine scenario. However, with data distributed training, we need to send the content in histogram buffer to other machines. With num_data <= 0, we exit the ConstructHistograms method directly, without clearing the buffer.Lines 1192 to 1193 in 6ad3e6e
So, when training a second tree, the empty leaf (though the leaf is actually not empty, considering the global training data) in a local machine will send the histogram content from the first iteration to other machines, which results in an incorrect global histogram. That's why the problem always occurs from a second iteration. Let me open a PR to fix this. |
I believe this is just a logging bug (bad wording):
|
|
oooo ok I'll submit a PR to fix that |
|
@jmoralez are you seeing output like this from lightgbm: [LightGBM] [Debug] Trained a tree with leaves = 31 and max_depth = 13 I'm seeing this for the regressor, where the last tree seems to output very high values The image you had above where split gain is NaN seems to resonate with what I am seeing, but it seems unlike mmlspark code dask code continue to call update, it ignores the value it returns: https://github.com/microsoft/LightGBM/blob/master/python-package/lightgbm/dask.py#L90 I created an issue here: |
|
@imatiach-msft Not sure whether the two are related. Which distributed training strategy are you using? |
|
@shiyu1994 it's using data_parallel training strategy |
|
Fixed by #4185 |
|
Hi @shiyu1994. Is it possible that this could still be present when splitting categorical features? I think I'm experiencing the same issue when I use them. |
|
@jose-moralez are you sure you are using the latest lightgbm on master? This issue was only recently fixed. If so, I would suggest creating a new issue. A similar issue a customer had in mmlspark was resolved with the new change. |
|
You're right @imatiach-msft, I had installed from source and checked that the issue was solved but I recently rebuilt my env and installed |
|
@jose-moralez ah ok, great to hear it's resolved then - if you still see issues though I would either just reopen this one or create a new one, it's difficult to keep track of closed issues (at least for me when I work on other open source projects as a maintainer) |
|
This issue has been automatically locked since there has not been any recent activity since it was closed. To start a new related discussion, open a new issue at https://github.com/microsoft/LightGBM/issues including a reference to this. |







Description
Hi. I'm trying to use LightGBM in time series forecasting and it works fine with local models. However, when using dask the predictions sometimes are huge (my time series are in the range [0, 160] and the predictions are 10^38). I've included a reproducible example (I'm sorry it's not that minimal but I couldn't reproduce it with some regular datasets). By running the example below sometimes the values are kind of big but other times they just really explode. With bigger datasets I've seen that the first tree is pretty much the same between local and dask but by the second tree the values in dask become increasingly large (so it may have something to do with the gradients). I haven't been able to track down the issue and was hoping someone could point me in the right direction.
Reproducible example
Environment info
LightGBM version or commit hash: 6356e65
Command(s) you used to install LightGBM
git clone --recursive https://github.com/microsoft/LightGBM.git cd LightGBM/python-package python setup.py installHere are some example plots.
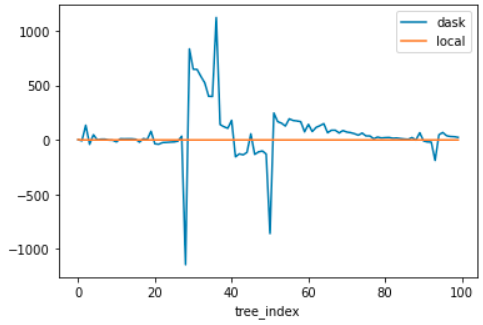
Sometimes the values are just "kind of big".
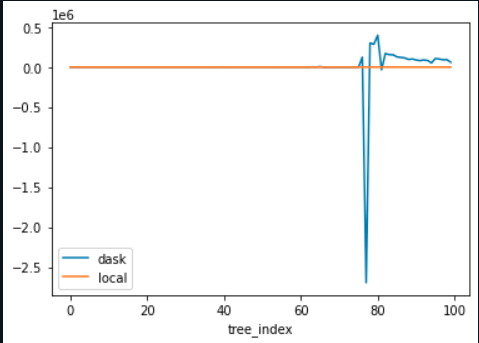
But other times they just really explode.
The text was updated successfully, but these errors were encountered: