-
Notifications
You must be signed in to change notification settings - Fork 24
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
4 changed files
with
53 additions
and
2 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,48 @@ | ||
| # GES algorithm: Basic examples | ||
|
|
||
| Causal discovery with the GES algorithm goes along the same lines as the PC algorithm. We again take the examle in chapter 2 of Judea Pearl's book. The causal model we are going to study can be represented using the following DAG: | ||
|
|
||
| 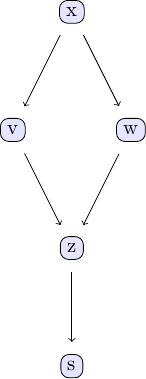 | ||
|
|
||
| We can easily create some sample data that corresponds to the causal structure described by the DAG. For the sake of simplicity, let's create data from a simple linear model that follows the structure defined by the DAG shown above: | ||
|
|
||
| ```Julia | ||
| using CausalInference | ||
| using TikzGraphs | ||
| using Random | ||
| Random.seed!(1) | ||
|
|
||
| # Generate some sample data to use with the GES algorithm | ||
|
|
||
| N = 2000 # number of data points | ||
|
|
||
| # define simple linear model with added noise | ||
| x = randn(N) | ||
| v = x + randn(N)*0.25 | ||
| w = x + randn(N)*0.25 | ||
| z = v + w + randn(N)*0.25 | ||
| s = z + randn(N)*0.25 | ||
|
|
||
| df = (x=x, v=v, w=w, z=z, s=s) | ||
| ``` | ||
|
|
||
| With this data ready, we can now see to what extent we can back out the underlying causal structure from the data using the GES algorithm. Under the hood, GES uses a score to determine the causal relationships between different variables in a given data set. By default, `ges` uses a Gaussian BIC to score different causal models. | ||
|
|
||
| ```Julia | ||
| est_g, score = ges(df; penalty=1.0, parallel=true) | ||
| ``` | ||
|
|
||
| In order to investigate the output of the GES algorithm, the same plotting function applies. | ||
|
|
||
| ```Julia | ||
| tp = plot_pc_graph_tikz(est_g, [String(k) for k in keys(df)]) | ||
| ``` | ||
|
|
||
| 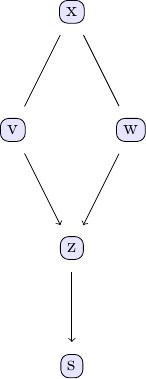 | ||
|
|
||
| We can compare with the output of the PC algorithm | ||
| ``` | ||
| est_g2 = pcalg(df, 0.01, gausscitest) | ||
| est_g == est_g2 | ||
| ``` | ||
| For the data using this seed, both agree. |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters