Fast non-linear gravity inversion in spherical coordinates with application to the South American Moho
by Leonardo Uieda and Valéria C. F. Barbosa
Published in the Geophysical Journal International: doi:10.1093/gji/ggw390
Click on this button to run the code online:
An archived snapshot of this repository is available on figshare at doi:10.6084/m9.figshare.3987267 (the manuscript LaTeX sources are not included).
A PDF of the article is available at leouieda.com/papers/paper-moho-inversion-tesseroids-2016.html
Contents:
- Python source code that implements the Moho inversion method proposed in the
paper (see
code/mohoinv.py). The modeling code uses the open-source geophysics library Fatiando a Terra. - Data files required to produce all results and figures presented in the paper.
- Source code in Jupyter notebooks that produce all
results and figures (the
.ipynbfiles incode). You can view static (not executable) versions of these notebooks through the links provided incode/README.md. - All figures in EPS format for the paper.
- Latex source to produce the manuscript (not included in the figshare archive).
- The estimated Moho depth model for South America, available in the file
model/south-american-moho.txtin ASCII xyz format (download the file).
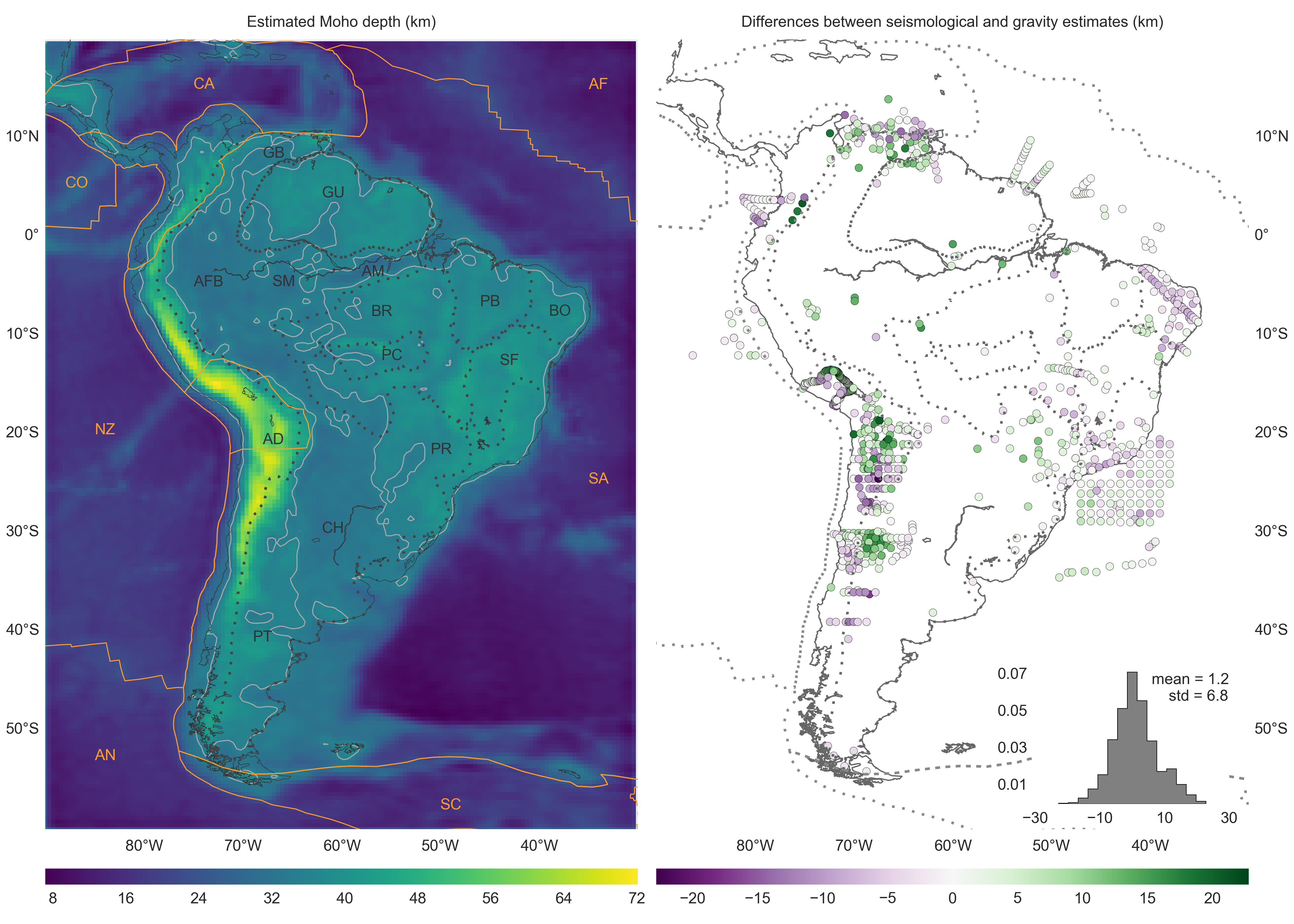
The main result from this publication is the gravity-derived Moho depth model for South America and the differences between it and seismological estimates of Assumpção et al. (2013):
Dotted lines represent the boundaries between major geologic provinces. AD: Andean Province, AFB: Andean foreland basins, AM: Amazonas Basin, BR: Brazilian Shield, BO: Borborema province, CH: Chaco Basin, GB: Guyana Basin, GU: Guyana Shield, PB: Parnaíba Basin, PC: Parecis Basin, PR: Paraná Basin, PT: Patagonia province, SF: São Francisco Craton, SM: Solimões Basin. Solid orange lines mark the limits of the main lithospheric plates. AF: Africa Plate, AN: Antarctica Plate, CA: Caribbean Plate, CO: Cocos Plate, SA: South America Plate, SC: Scotia Plate, NZ: Nazca Plate. The solid light grey line is the 35 km Moho depth contour.
Estimating the relief of the Moho from gravity data is a computationally intensive non-linear inverse problem. What is more, the modeling must take the Earths curvature into account when the study area is of regional scale or greater. We present a regularized non-linear gravity inversion method that has a low computational footprint and employs a spherical Earth approximation. To achieve this, we combine the highly efficient Bott's method with smoothness regularization and a discretization of the anomalous Moho into tesseroids (spherical prisms). The computational efficiency of our method is attained by harnessing the fact that all matrices involved are sparse. The inversion results are controlled by three hyper-parameters: the regularization parameter, the anomalous Moho density-contrast, and the reference Moho depth. We estimate the regularization parameter using the method of hold-out cross-validation. Additionally, we estimate the density-contrast and the reference depth using knowledge of the Moho depth at certain points. We apply the proposed method to estimate the Moho depth for the South American continent using satellite gravity data and seismological data. The final Moho model is in accordance with previous gravity-derived models and seismological data. The misfit to the gravity and seismological data is worse in the Andes and best in oceanic areas, central Brazil and Patagonia, and along the Atlantic coast. Similarly to previous results, the model suggests a thinner crust of 30-35 km under the Andean foreland basins. Discrepancies with the seismological data are greatest in the Guyana Shield, the central Solimões and Amazonas Basins, the Paraná Basin, and the Borborema province. These differences suggest the existence of crustal or mantle density anomalies that were unaccounted for during gravity data processing.
You can run the Jupyter notebooks online without installing anything through the free Binder webservice:
mybinder.org/repo/pinga-lab/paper-moho-inversion-tesseroids
Start by navigating to the code folder and choose a notebook to execute.
Beware that the CRUST1.0 and South American Moho notebooks will probably not
be able to run on Binder because they take too long (a few days).
If you want to try out the method start with the code/synthetic-simple.ipynb
notebook or see how the figures for the paper are generated in the
code/paper-figures.ipynb notebook.
Tip: use Shift + Enter to execute a code cell.
If you want to run the full inversion on your machine, follow the steps below.
First, download a copy of all files in this repository:
-
Or clone the git repository:
git clone https://github.com/pinga-lab/paper-moho-inversion-tesseroids.git
You'll need a working Python 2.7 environment with all the standard scientific packages installed (numpy, scipy, matplotlib, etc). The easiest (and recommended) way to get this is to download and install the Anaconda Python distribution. Make sure you get the Python 2.7 version.
All required dependencies are specified in the environmet.yml file.
The exact versions of every dependency are also specified in the conda-*.lock
files. The lock files can be used to create an exact version of the environment
used in the paper for Linux, Mac, and Windows.
You can use conda package manager (included in Anaconda) to create a virtual
environment with all the required packages installed. First, unzip the contents
of this repository (if you've downloaded the zip file) and cd into the root
of the repository (where environment.yml and the conda-*.lock files are
located). Next, run the following command in a terminal (substituting the
appropriate lock file for your operating system):
conda create -n moho --file conda-linux-64.lock
This will create the virtual environment based on the specifications in
environment.yml and install all required dependencies.
To activate the conda environment, run
source activate moho
or, if you're on Windows,
activate moho
This will enable the environment for your current terminal session.
The inversion and figure generation are all run inside Jupyter notebooks. To
execute them, you must first start the notebook server by going into the
repository folder and running (make sure you have the conda environment
enabled first):
jupyter notebook
This will start the server and open your default web browser to the Jupyter
console interface. In the page, go into the code folder and select the
notebook that you wish to view/run.
Here what a notebook looks like running in Firefox:
The notebook is divided cells (some have text while other have code). Each can
be executed using Shift + Enter. Executing text cells does nothing and
executing code cells runs the code and produces it's output. To execute the
whole notebook, run all cells in order.
All results figures in the paper are generated by the
code/paper-figures.ipynb notebook.
All source code is made available under a BSD 3-clause license. You can freely
use and modify the code, without warranty, so long as you provide attribution
to the authors. See LICENSE.md for the full license text.
The model results in file model/south-american-moho.txt are available under
the Creative Commons Attribution 4.0 License (CC-BY).
The manuscript text is not open source. The authors reserve the rights to the article content, which has been accepted for publication in the Geophysical Journal International.