Jenetics is an Genetic Algorithm, Evolutionary Algorithm and Genetic Programming library, respectively, written in Java. It is designed with a clear separation of the several concepts of the algorithm, e.g. Gene, Chromosome, Genotype, Phenotype, Population and fitness Function. Jenetics allows you to minimize and maximize the given fitness function without tweaking it. In contrast to other GA implementations, the library uses the concept of an evolution stream (EvolutionStream) for executing the evolution steps. Since the EvolutionStream implements the Java Stream interface, it works smoothly with the rest of the Java Stream API.
Other languages
- Jenetics.Net: Experimental .NET Core port in C# of the base library.
- Helisa: Scala wrapper around the Jenetics library.
The library is fully documented (javadoc) and comes with an user manual (pdf).
- JRE 8: Java runtime version 8 is needed for using the library, respectively for running the examples.
- JDK 8: The Java JDK 8 must be installed.
- Gradle 5.x: Gradle is used for building the library. (Gradle is download automatically, if you are using the Gradle Wrapper script
./gradlew, located in the base directory, for building the library.)
- TestNG 6.x: Jenetics uses TestNG framework for unit tests.
- Apache Commons Math 3.6: Library is used for testing statistical collectors.
For building the Jenetics library from source, download the most recent, stable package version from Github and extract it to some build directory.
$ unzip jenetics-<version>.zip -d <builddir>
<version> denotes the actual Jenetics version and <builddir> the actual build directory. Alternatively you can check out the master branch from Github.
$ git clone https://github.com/jenetics/jenetics.git <builddir>
Jenetics uses Gradle as build system and organizes the source into sub-projects (modules). Each sub-project is located in it’s own sub-directory:
Published projects
The following projects/modules are also published to Maven.
- jenetics
: This project contains the source code and tests for the Jenetics core-module.
- jenetics.ext
: This module contains additional non-standard GA operations and data types. It also contains classes for solving multi-objective problems (MOEA).
- jenetics.prog
: The modules contains classes which allows to do genetic programming (GP). It seamlessly works with the existing
EvolutionStreamand evolutionEngine. - jenetics.xml
: XML marshalling module for the Jenetics base data structures.
Non-published projects
- jenetics.example: This project contains example code for the core-module.
- jenetics.doc: Contains the code of the web-site and the manual.
- jenetics.tool: This module contains classes used for doing integration testing and algorithmic performance testing. It is also used for creating GA performance measures and creating diagrams from the performance measures.
For building the library change into the <builddir> directory (or one of the module directory) and call one of the available tasks:
- compileJava: Compiles the Jenetics sources and copies the class files to the
<builddir>/<module-dir>/build/classes/maindirectory. - jar: Compiles the sources and creates the JAR files. The artifacts are copied to the
<builddir>/<module-dir>/build/libsdirectory. - javadoc: Generates the API documentation. The Javadoc is stored in the
<builddir>/<module-dir>/build/docsdirectory - test: Compiles and executes the unit tests. The test results are printed onto the console and a test-report, created by TestNG, is written to
<builddir>/<module-dir>directory. - clean: Deletes the
<builddir>/build/*directories and removes all generated artifacts.
For building the library jar from the source call
$ cd <build-dir>
$ ./gradlew jar
The minimum evolution Engine setup needs a genotype factory, Factory<Genotype<?>>, and a fitness Function. The Genotype implements the Factory interface and can therefore be used as prototype for creating the initial Population and for creating new random Genotypes.
import io.jenetics.BitChromosome;
import io.jenetics.BitGene;
import io.jenetics.Genotype;
import io.jenetics.engine.Engine;
import io.jenetics.engine.EvolutionResult;
import io.jenetics.util.Factory;
public class HelloWorld {
// 2.) Definition of the fitness function.
private static Integer eval(Genotype<BitGene> gt) {
return gt.getChromosome()
.as(BitChromosome.class)
.bitCount();
}
public static void main(String[] args) {
// 1.) Define the genotype (factory) suitable
// for the problem.
Factory<Genotype<BitGene>> gtf =
Genotype.of(BitChromosome.of(10, 0.5));
// 3.) Create the execution environment.
Engine<BitGene, Integer> engine = Engine
.builder(HelloWorld::eval, gtf)
.build();
// 4.) Start the execution (evolution) and
// collect the result.
Genotype<BitGene> result = engine.stream()
.limit(100)
.collect(EvolutionResult.toBestGenotype());
System.out.println("Hello World:\n" + result);
}
}In contrast to other GA implementations, the library uses the concept of an evolution stream (EvolutionStream) for executing the evolution steps. Since the EvolutionStream implements the Java Stream interface, it works smoothly with the rest of the Java streaming API. Now let's have a closer look at listing above and discuss this simple program step by step:
-
The probably most challenging part, when setting up a new evolution
Engine, is to transform the problem domain into a appropriateGenotype(factory) representation. In our example we want to count the number of ones of aBitChromosome. Since we are counting only the ones of one chromosome, we are adding only oneBitChromosometo ourGenotype. In general, theGenotypecan be created with 1 to n chromosomes. -
Once this is done, the fitness function which should be maximized, can be defined. Utilizing the new language features introduced in Java 8, we simply write a private static method, which takes the genotype we defined and calculate it's fitness value. If we want to use the optimized bit-counting method,
bitCount(), we have to cast theChromosome<BitGene>class to the actual usedBitChromosomeclass. Since we know for sure that we created the Genotype with aBitChromosome, this can be done safely. A reference to the eval method is then used as fitness function and passed to theEngine.buildmethod. -
In the third step we are creating the evolution
Engine, which is responsible for changing, respectively evolving, a given population. TheEngineis highly configurable and takes parameters for controlling the evolutionary and the computational environment. For changing the evolutionary behavior, you can set different alterers and selectors. By changing the usedExecutorservice, you control the number of threads, the Engine is allowed to use. An newEngineinstance can only be created via its builder, which is created by calling theEngine.buildermethod. -
In the last step, we can create a new
EvolutionStreamfrom ourEngine. TheEvolutionStreamis the model or view of the evolutionary process. It serves as a »process handle« and also allows you, among other things, to control the termination of the evolution. In our example, we simply truncate the stream after 100 generations. If you don't limit the stream, theEvolutionStreamwill not terminate and run forever. Since theEvolutionStreamextends thejava.util.stream.Streaminterface, it integrates smoothly with the rest of the Java Stream API. The final result, the bestGenotypein our example, is then collected with one of the predefined collectors of theEvolutionResultclass.
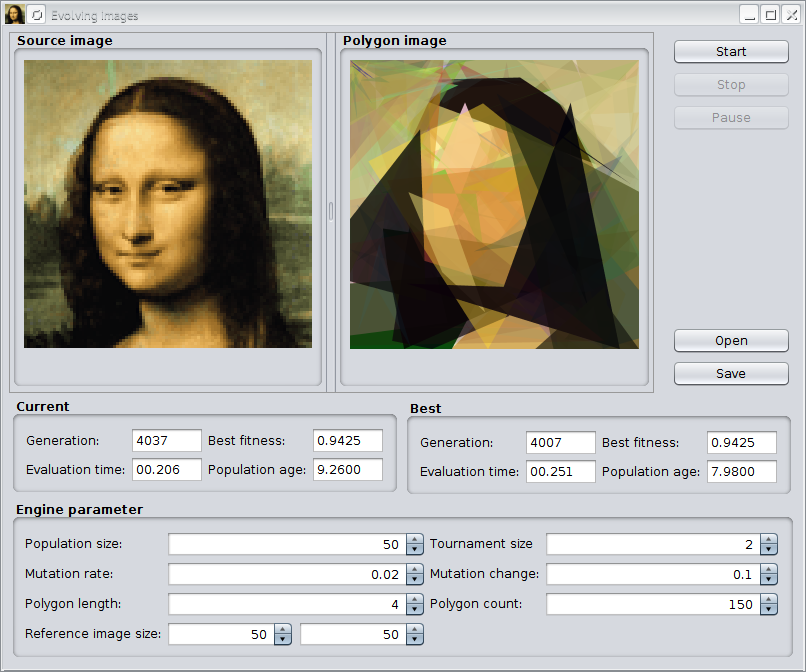
This example tries to approximate a given image by semitransparent polygons. It comes with an Swing UI, where you can immediately start your own experiments. After compiling the sources with
$ ./gradlew compileTestJava
you can start the example by calling
$ ./jrun io.jenetics.example.image.EvolvingImages
The previous image shows the GUI after evolving the default image for about 4,000 generations. With the »Open« button it is possible to load other images for polygonization. The »Save« button allows to store polygonized images in PNG format to disk. At the button of the UI, you can change some of the GA parameters of the example.
- Renaissance Suite: Renaissance is a modern, open, and diversified benchmark suite for the JVM, aimed at testing JIT compilers, garbage collectors, profilers, analyzers and other tools.
- Chartsy|One: Chartsy|One is a Netbeans based tool for stock market investors and traders.
- Chronetic: Chronetic is an open-source time pattern analysis library built to describe time-series data.
- APP4MC: Eclipse APP4MC is a platform for engineering embedded multi- and many-core software systems.
- Introduction to Jenetics Library, by baeldung, April 11. 2017
- How to Solve Tough Problems Using Genetic Algorithms, by Tzofia Shiftan, April 6. 2017
- Genetic algorithms with Java, by William Antônio, January 10. 2017
- Jenetics 설치 및 예제, by JDM, May 8. 2015
- 유전 알고리즘 (Genetic Algorithms), by JDM, April 2. 2015
- Maxime Cordy, Steve Muller, Mike Papadakis, and Yves Le Traon. Search-based test and improvement of machine-learning-based anomaly detection systems. Proceedings of the 28th ACM SIGSOFT International Symposium on Software Testing and Analysis (ISSTA 2019). ACM, New York, NY, USA, 158-168. July 2019.
- Michael Trotter, Timothy Wood and Jinho Hwang. Forecasting a Storm: Divining Optimal Configurations using Genetic Algorithms and Supervised Learning. 13th IEEE International Conference on Self-Adaptive and Self-Organizing Systems (SASO 2019). June 2019.
- Krawczyk, Lukas & Bazzal, Mahmoud & Prasath Govindarajan, Ram & Wolff, Carsten. An analytical approach for calculating end-to-end response times in autonomous driving applications. 10th International Workshop on Analysis Tools and Methodologies for Embedded and Real-time Systems (WATERS 2019). June 2019.
- Rodolfo Ayala Lopes, Thiago Macedo Gomes, and Alan Robert Resende de Freitas. A symbolic evolutionary algorithm software platform. Proceedings of the Genetic and Evolutionary Computation Conference Companion (GECCO '19). July 2019.
- Aleksandar Prokopec, Andrea Rosà, David Leopoldseder, Gilles Duboscq, Petr Tůma, Martin Studener, Lubomír Bulej, Yudi Zheng, Alex Villazón, Doug Simon, Thomas Würthinger, Walter Binder. Renaissance: Benchmarking Suite for Parallel Applications on the JVM. PLDI ’19, Phoenix, AZ, USA. June 2019.
- Robert Höttger, Lukas Krawczyk, Burkhard Igel, Olaf Spinczyk. Memory Mapping Analysis for Automotive Systems. Brief Presentations Proceedings (RTAS 2019). Apr. 2019.
- Al Akkad, M. A., & Gazimzyanov, F. F. AUTOMATED SYSTEM FOR EVALUATING 2D-IMAGE COMPOSITIONAL CHARACTERISTICS: CONFIGURING THE MATHEMATICAL MODEL. Intellekt. Sist. Proizv., 17(1), 26-33. doi: 10.22213/2410-9304-2019-1-26-33. Apr. 2019.
- Alcayde, A.; Baños, R.; Arrabal-Campos, F.M.; Montoya, F.G. Optimization of the Contracted Electric Power by Means of Genetic Algorithms. Energies, Volume 12, Issue 7, Apr. 2019.
- Aleksandar Prokopec, Andrea Rosà, David Leopoldseder, Gilles Duboscq, Petr Tůma, Martin Studener, Lubomír Bulej, Yudi Zheng, Alex Villazón, Doug Simon, Thomas Wuerthinger, Walter Binder. On Evaluating the Renaissance Benchmarking Suite: Variety, Performance, and Complexity. Cornell University: Programming Languages, Mar. 2019.
- S. Appel, W. Geithner, S. Reimann, M Sapinski, R. Singh, D. M. Vilsmeier OPTIMIZATION OF HEAVY-ION SYNCHROTRONS USINGNATURE-INSPIRED ALGORITHMS AND MACHINE LEARNING.13th Int. Computational Accelerator Physics Conf., Feb. 2019.
- Saad, Christian, Bernhard Bauer, Ulrich R Mansmann, and Jian Li. AutoAnalyze in Systems Biology. Bioinformatics and Biology Insights, Jan. 2019.
- Gandeva Bayu Satrya, Soo Young Shin. Evolutionary Computing Approach to Optimize Superframe Scheduling on Industrial Wireless Sensor Networks. Cornell University, Dec. 2018.
- H.R. Maier, S. Razavi, Z. Kapelan, L.S. Matott, J. Kasprzyk, B.A. Tolson. Introductory overview: Optimization using evolutionary algorithms and other metaheuristics. Environmental Modelling & Software, Dec. 2018.
- Erich C. Teppan and Giacomo Da Col. Automatic Generation of Dispatching Rules for Large Job Shops by Means of Genetic Algorithms. CIMA 2018, International Workshop on Combinations of Intelligent Methods and Applications, Nov. 2018.
- Pasquale Salzaa, Filomena Ferrucci. Speed up genetic algorithms in the cloud using software containers. Future Generation Computer Systems, Oct. 2018.
- Ghulam Mubashar Hassan and Mark Reynolds. Genetic Algorithms for Scheduling and Optimization of Ore Train Networks. GCAI-2018. 4th Global Conference on Artificial Intelligence, Sep. 2018.
- Drezewski, Rafal & Kruk, Sylwia & Makowka, Maciej. The Evolutionary Optimization of a Company’s Return on Equity Factor: Towards the Agent-Based Bio-Inspired System Supporting Corporate Finance Decisions. IEEE Access. 6. 10.1109/ACCESS.2018.2870201, Sep. 2018.
- Arifin, H. H., Chimplee, N. , Kit Robert Ong, H. , Daengdej, J. and Sortrakul, T. Automated Component‐Selection of Design Synthesis for Physical Architecture with Model‐Based Systems Engineering using Evolutionary Trade‐off. INCOSE International Symposium, 28: 1296-1310, Aug. 2018.
- Stephan Pirnbaum. Die Evolution im Algorithmus - Teil 2: Multikriterielle Optimierung und Architekturerkennung. JavaSPEKTRUM 03/2018, pp 66–69, May 2018.
- W. Geithner, Z. Andelkovic, S. Appel, O. Geithner, F. Herfurth, S. Reimann, G. Vorobjev, F. Wilhelmstötter. Genetic Algorithms for Machine Optimization in the Fair Control System Environment. The 9th International Particle Accelerator Conference (IPAC'18), May 2018.
- Stephan Pirnbaum. Die Evolution im Algorithmus - Teil 1: Grundlagen. JavaSPEKTRUM 01/2018, pp 64–68, Jan. 2018.
- Alexander Felfernig, Rouven Walter, José A. Galindo, David Benavides, Seda Polat Erdeniz, Müslüm Atas, Stefan Reiterer. Anytime diagnosis for reconfiguration. Journal of Intelligent Information Systems, pp 1–22, Jan. 2018.
- Bruce A. Johnson. From Raw Data to Protein Backbone Chemical Shifts Using NMRFx Processing and NMRViewJ Analysis. Protein NMR: Methods and Protocols, pp. 257--310, Springer New York, Nov. 2017.
- Cuadra P., Krawczyk L., Höttger R., Heisig P., Wolff C. Automated Scheduling for Tightly-Coupled Embedded Multi-core Systems Using Hybrid Genetic Algorithms. Information and Software Technologies: 23rd International Conference, ICIST 2017, Druskininkai, Lithuania. Communications in Computer and Information Science, vol 756. Springer, Cham, Sep. 2017.
- Michael Trotter, Guyue Liu, Timothy Wood. Into the Storm: Descrying Optimal Configurations Using Genetic Algorithms and Bayesian Optimization. Foundations and Applications of Self* Systems (FAS*W), 2017 IEEE 2nd International Workshops Sep. 2017.
- Emna Hachicha, Karn Yongsiriwit, Mohamed Sellami. Genetic-Based Configurable Cloud Resource Allocation in QoS-Aware Business Process Development. Information and Software Technologies: 23rd International Conference, ICIST 2017, Druskininkai, Lithuania. Web Services (ICWS), 2017 IEEE International Conference, Jun. 2017.
- Abraão G. Nazário, Fábio R. A. Silva, Raimundo Teive, Leonardo Villa, Antônio Flávio, João Zico, Eire Fragoso, Ederson F. Souza. Automação Domótica Simulada Utilizando Algoritmo Genético Especializado na Redução do Consumo de Energia. Computer on the Beach 2017 pp. 180-189, March 2017.
- Bandaru, S. and Deb, K. Metaheuristic Techniques. Decision Sciences. CRC Press, pp. 693-750, Nov. 2016.
- Lyazid Toumi, Abdelouahab Moussaoui, and Ahmet Ugur. EMeD-Part: An Efficient Methodology for Horizontal Partitioning in Data Warehouses. Proceedings of the International Conference on Intelligent Information Processing, Security and Advanced Communication. Djallel Eddine Boubiche, Faouzi Hidoussi, and Homero Toral Cruz (Eds.). ACM, New York, NY, USA, Article 43, 7 pages, 2015.
- Andreas Holzinger (Editor), Igo Jurisica (Editor). Interactive Knowledge Discovery and Data Mining in Biomedical Informatics. Lecture Notes in Computer Science, Vol. 8401. Springer, 2014.
- Lyazid Toumi, Abdelouahab Moussaoui, Ahmet Ugur. Particle swarm optimization for bitmap join indexes selection problem in data warehouses. The Journal of Supercomputing, Volume 68, Issue 2, pp 672-708, May 2014.
- TANG Yi (Guangzhou Power Supply Bureau Limited, Guangzhou 511400, China) Study on Object-Oriented Reactive Compensation Allocation Optimization Algorithm for Distribution Networks, Oct. 2012.
- John M. Linebarger, Richard J. Detry, Robert J. Glass, Walter E. Beyeler, Arlo L. Ames, Patrick D. Finley, S. Louise Maffitt. Complex Adaptive Systems of Systems Engineering Environment Version 1.0. SAND REPORT, Feb. 2012.
- #550: Erroneous index check for
Sample.argAt(int)method inio.jenetics.prog.regressionpackage. - #554:
ClassCastExceptioninio.jenetics.prog.regression.Regressionclass.
- #534: Generify
Regressionclasses so it can be used for regression analysis of arbitrary types. - #529: Implementation of Hybridizing PSM and RSM mutation operator (HPRM)
- #518: Implementation of Symbolic Regression classes. This makes it easier to solve such optimization problems.
- #515: Rename
Tree.getIndex(Tree)toTree.indexOf(Tree). - #509: Allow to collect the nth best optimization results.
final ISeq<EvolutionResult<DoubleGene, Double>> best = engine.stream()
.limit(Limits.bySteadyFitness(50))
.flatMap(MinMax.toStrictlyIncreasing())
.collect(ISeq.toISeq(10));- #504: Rename
Tree.getChild(int)toTree.childAt(int). - #500: Implementation of Reverse Sequence mutation operator (RSM).
- #497: Implement Boolean operators for GP.
- #496: Implement
GToperator for GP. - #493: Add dotty tree formatter
- #488: Implement new tree formatter
TreeFormatter.LISP. This allows to create a Lisp string representation from a givenTree. - #487: Re-implementation of 'MathTreePruneAlterer'. The new implementation uses the newly introduced Tree Rewriting API, implemented in #442.
- #486: Implement
TreeRewriteAlterer, based on the new Tree Rewriting API. - #485: Cleanup of
MathExprclass. - #484: The
Tree.toString()now returns a parentheses string. - #481: The parentheses tree representation now only escapes "protected" characters.
- #469: Implementation of additional
Evaluatorfactory methods. - #465: Remove fitness scaler classes. The fitness scaler doesn't carry its weight.
- #455: Implementation of
CompletableFutureEvaluator. - #450: Improvement of
FutureEvaluatorclass. - #449: The
Engine.Builderconstructor is now public and is the most generic way for creating engine builder instances. All other builder factory methods are calling this primary constructor. - #447: Remove evolution iterators. The whole evolution is no performed via streams.
- #442: Introduce Tree Rewriting API, which allows to define own rewrite rules/system. This is very helpful when solving GP related problems.
final TRS<String> trs = TRS.parse(
"add(0,$x) -> $x",
"add(S($x),$y) -> S(add($x,$y))",
"mul(0,$x) -> 0",
"mul(S($x),$y) -> add(mul($x,$y),$y)"
);
// Converting the input tree into its normal form.
final TreeNode<String> tree = TreeNode.parse("add(S(0),S(mul(S(0),S(S(0)))))");
trs.rewrite(tree);
assert tree.equals(TreeNode.parse("S(S(S(S(0))))"));- #372: Allow to define the chromosome index an
Altereris allowed to change. This allows to define alterers for specific chromosomes in a genotype.
// The genotype prototype, consisting of 4 chromosomes
final Genotype<DoubleGene> gtf = Genotype.of(
DoubleChromosome.of(0, 1),
DoubleChromosome.of(1, 2),
DoubleChromosome.of(2, 3),
DoubleChromosome.of(3, 4)
);
// Define the GA engine.
final Engine<DoubleGene, Double> engine = Engine
.builder(gt -> gt.getGene().doubleValue(), gtf)
.selector(new RouletteWheelSelector<>())
.alterers(
// The `Mutator` is used on chromosome with index 0 and 2.
PartialAlterer.of(new Mutator<DoubleGene, Double>(), 0, 2),
// The `MeanAlterer` is used on chromosome 3.
PartialAlterer.of(new MeanAlterer<DoubleGene, Double>(), 3),
// The `GaussianMutator` is used on all chromosomes.
new GaussianMutator<>()
)
.build();- #368: Remove deprecated code.
- #364: Clean implementation of async fitness functions.
- #342: The
Treeaccessor names are no longer in a Java Bean style:getChild(int)->childAt(int). This corresponds to thechildAtPath(path)methods. - #331: Remove
hashCodeandequalsmethod fromSelectorandAlterer. - #314: Add factory method for
AdaptiveEngine, which simplifies its creation. - #308: General improvement of object serialization.
- #50: Improve Genotype validation. The new
Constraintinterface, and its implementationRetryConstraint, now allows a finer control of the validation and recreation of individuals.
- #520: Fix tree-rewriting for
Constvalues. This leads to non-matching nodes when trying to simplify the GP tree. - #475: Level function returns different results depending on whether the iterator is iterating through a
ProgramGeneorTreeNode. - #473:
DynamicGenotypeexample causesIllegalArgumentException.
The library is licensed under the Apache License, Version 2.0.
Copyright 2007-2019 Franz Wilhelmstötter
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.