This repository contains code for training and running climate neural network surrogate models. For detais on various experiments visit our site https://stresearch.github.io/gaia/
Warning: This is an active research project. The code base is constantly evolving as new features are being added and old ones are depreciated.
This work is part of the DARPA ACTME (AI-assisted Climate Tipping-point Modeling) AIE Program - https://github.com/ACTM-darpa/info-and-links
- Installation
- Data Preprocessing
- Configuration Parameters
- Training
- Inference
- Generate Diagnostic Plots
- Export Model for Integration
- Pre-trained Models
Install requirments:
git clone https://github.com/stresearch/gaia
pip install -r requirementsWe provide a toy dataset here. It’s subsampled cam4 dataset.
To prerocess large scale exports from climate model runs. we work with
outputs from two climate models: CAM4 and SPCAM. - We assume raw data
resides in an S3 bucket with one file per day in the NCDF4 format. -
To prepocess the data we use a fairy large AWS EC instance: -
r4.16xlarge with 64 CPUs - attach at least 500GB EBS volume for
local caching
To run prepocessing from an AWS instance with default parameters for
split=train,test:
NCDataConstructor.default_data(
cls,
split="train",
bucket_name="name_of_bucket",
prefix="spcamclbm-nx-16-20m-timestep",
save_location=".",
train_years = 2,
cache = ".",
workers = 64
)We assume the following input/output variables:
This should generate 4 files:
spcamclbm-nx-16-20m-timestep_4_test.pt spcamclbm-nx-16-20m-timestep_4_val.pt spcamclbm-nx-16-20m-timestep_4_train.pt spcamclbm-nx-16-20m-timestep_4_var_index.pt
Copy to machine where you want to train the model. For more details see
`gaia.data
module <
Line 454 in c0268fa
To perform training, we use a machine with at least a single GPU and
64GBs of RAM (to load the full dataset into memory, smaller for a toy
dataset). To use the toy dataset, set the environmental variable
GAIA_TOY_DATA prefix where it is located.
Configure the data, model and training parameters. We specify
mode, dataset, inputs, outputs, batch_size, model_type, gpu and max-epochs
import sys
import os
import glob
from gaia.training import main
from gaia.config import Config
os.environ["GAIA_TOY_DATA"] = "/ssddg1/gaia/cam4_v5/cam4-famip-30m-timestep-third-upload"
inputs = ['B_Q [t+1]',
'B_T [t+1]',
'B_U [t+1]',
'B_V [t+1]',
'B_OMEGA [t+1]',
'B_Z3 [t+1]',
'B_PS [t+1]',
'SOLIN [t+1]',
'B_SHFLX [t+1]',
'B_LHFLX [t+1]',
'LANDFRAC [t]',
'OCNFRAC [t]',
'ICEFRAC [t]',
'FSNS [t]',
'FLNS [t]',
'FSNT [t]',
'FLNT [t]',
'FSDS [t]']
outputs = ['A_PTTEND [t+1]',
'A_PTEQ [t+1]',
'FSNS [t+1]',
'FLNS [t+1]',
'FSNT [t+1]',
'FLNT [t+1]',
'FSDS [t+1]',
'FLDS [t+1]',
'SRFRAD [t+1]',
'SOLL [t+1]',
'SOLS [t+1]',
'SOLLD [t+1]',
'SOLSD [t+1]',
'PRECT [t+1]',
'PRECC [t+1]',
'PRECL [t+1]',
'PRECSC [t+1]',
'PRECSL [t+1]']
config = Config(
{
"mode": "train,test,predict",
"dataset_params": {
"dataset": "toy",
"inputs": inputs,
"outputs": outputs,
"batch_size": 4096,
},
"trainer_params": {"gpus": [gpu], "max_epochs": 100},
"model_params": {
"model_type": "fcn",
},
}
)This is what the full config file looks.
print(config)
dataset_params:
batch_size: 4096
dataset: cam4_toy
inputs:
- B_Q [t+1]
- B_T [t+1]
- B_U [t+1]
- B_V [t+1]
- B_OMEGA [t+1]
- B_Z3 [t+1]
- B_PS [t+1]
- SOLIN [t+1]
- B_SHFLX [t+1]
- B_LHFLX [t+1]
- LANDFRAC [t]
- OCNFRAC [t]
- ICEFRAC [t]
- FSNS [t]
- FLNS [t]
- FSNT [t]
- FLNT [t]
- FSDS [t]
mean_thres: 1.0e-13
outputs:
- A_PTTEND [t+1]
- A_PTEQ [t+1]
- FSNS [t+1]
- FLNS [t+1]
- FSNT [t+1]
- FLNT [t+1]
- FSDS [t+1]
- FLDS [t+1]
- SRFRAD [t+1]
- SOLL [t+1]
- SOLS [t+1]
- SOLLD [t+1]
- SOLSD [t+1]
- PRECT [t+1]
- PRECC [t+1]
- PRECL [t+1]
- PRECSC [t+1]
- PRECSL [t+1]
test:
batch_size: 4096
data_grid: &id001
- 3.5446380000000097
- 7.3888135000000075
- 13.967214000000006
- 23.944625
- 37.23029000000011
- 53.1146050000002
- 70.05915000000029
- 85.43911500000031
- 100.51469500000029
- 118.25033500000026
- 139.11539500000046
- 163.66207000000043
- 192.53993500000033
- 226.51326500000036
- 266.4811550000001
- 313.5012650000006
- 368.81798000000157
- 433.8952250000011
- 510.45525500000167
- 600.5242000000027
- 696.7962900000033
- 787.7020600000026
- 867.1607600000013
- 929.6488750000024
- 970.5548300000014
- 992.5560999999998
dataset_file: /ssddg1/gaia/cam4_v5/cam4-famip-30m-timestep-third-upload_test.pt
flatten: true
include_index: false
inputs: &id002
- B_Q [t+1]
- B_T [t+1]
- B_U [t+1]
- B_V [t+1]
- B_OMEGA [t+1]
- B_Z3 [t+1]
- B_PS [t+1]
- SOLIN [t+1]
- B_SHFLX [t+1]
- B_LHFLX [t+1]
- LANDFRAC [t]
- OCNFRAC [t]
- ICEFRAC [t]
- FSNS [t]
- FLNS [t]
- FSNT [t]
- FLNT [t]
- FSDS [t]
outputs: &id003
- A_PTTEND [t+1]
- A_PTEQ [t+1]
- FSNS [t+1]
- FLNS [t+1]
- FSNT [t+1]
- FLNT [t+1]
- FSDS [t+1]
- FLDS [t+1]
- SRFRAD [t+1]
- SOLL [t+1]
- SOLS [t+1]
- SOLLD [t+1]
- SOLSD [t+1]
- PRECT [t+1]
- PRECC [t+1]
- PRECL [t+1]
- PRECSC [t+1]
- PRECSL [t+1]
shuffle: false
space_filter: null
subsample: 1
subsample_mode: random
var_index_file: /ssddg1/gaia/cam4_v5/cam4-famip-30m-timestep-third-upload_var_index.pt
train:
batch_size: 4096
data_grid: *id001
dataset_file: /ssddg1/gaia/cam4_v5/cam4-famip-30m-timestep-third-upload_train.pt
flatten: false
include_index: false
inputs: *id002
outputs: *id003
shuffle: true
space_filter: null
subsample: 1
subsample_mode: random
var_index_file: /ssddg1/gaia/cam4_v5/cam4-famip-30m-timestep-third-upload_var_index.pt
val:
batch_size: 4096
data_grid: *id001
dataset_file: /ssddg1/gaia/cam4_v5/cam4-famip-30m-timestep-third-upload_val.pt
flatten: false
include_index: false
inputs: *id002
outputs: *id003
shuffle: false
space_filter: null
subsample: 1
subsample_mode: random
var_index_file: /ssddg1/gaia/cam4_v5/cam4-famip-30m-timestep-third-upload_var_index.pt
mode: train,test,predict
model_params:
ckpt: null
lr: 0.001
lr_schedule: cosine
model_config:
dropout: 0.01
hidden_size: 512
leaky_relu: 0.15
model_type: fcn
num_layers: 7
model_type: fcn
replace_std_with_range: false
use_output_scaling: false
weight_decay: 0
seed: true
trainer_params:
gpus:
- 5
max_epochs: 100
precision: 16For default parameters consult gaia.config.Config class. There are
three groups of parameters:
trainer_params, dataset_params, model_params .
Parameters can be specified by - directly passing nested dictionaries
for each - pass in nothing which will automatically read in defaults
from Config - command line arguments using the dot notation to
override specified Config defaults
Example configs:
dataset_params =
{'test': {'batch_size': 138240,
'dataset_file': '/ssddg1/gaia/cam4/cam4-famip-30m-timestep_4_test.pt',
'flatten': True,
'shuffle': False,
'var_index_file': '/ssddg1/gaia/cam4/cam4-famip-30m-timestep_4_var_index.pt'},
'train': {'batch_size': 138240,
'dataset_file': '/ssddg1/gaia/cam4/cam4-famip-30m-timestep_4_train.pt',
'flatten': False,
'shuffle': True,
'var_index_file': '/ssddg1/gaia/cam4/cam4-famip-30m-timestep_4_var_index.pt'},
'val': {'batch_size': 138240,
'dataset_file': '/ssddg1/gaia/cam4/cam4-famip-30m-timestep_4_val.pt',
'flatten': False,
'shuffle': False,
'var_index_file': '/ssddg1/gaia/cam4/cam4-famip-30m-timestep_4_var_index.pt'}}training_params =
{'precision': 16, 'max_epochs': 200, gpus=[0]}model_params =
{'lr': 0.001,
'optimizer': 'adam',
'model_config': {'model_type': 'fcn', 'num_layers': 7}}We support the following types of NN models:
fcn: baseline MLP
model_config = {
"model_type": "fcn",
"num_layers": 7,
"hidden_size": 512,
"dropout": 0.01,
"leaky_relu": 0.15
}fcn_history: baseline MLP with an extra input of memory variables i.e. outputs from previous time step
model_config = {
"model_type": "fcn_history",
"num_layers": 7,
"hidden_size": 512,
"leaky_relu": 0.15
}conv1d: same as fcn functionally but accepts an “image” like data i.e. image of lat,lon,variablles
model_config = {
"model_type": "conv1d",
"num_layers": 7,
"hidden_size": 128
}resdnn: architecture from [ref]
model_config = {
"model_type": "resdnn",
"num_layers": 7,
"hidden_size": 512,
"dropout": 0.01,
"leaky_relu": 0.15
}encoderdecoder: encoder/decoder with a bottleneck feature
model_config = {
"model_type": "encoderdecoder",
"num_layers": 7,
"hidden_size": 512,
"dropout": 0.01,
"leaky_relu": 0.15,
"bottleneck_dim": 32,
}transformer: transformer with z level positional encoding
model_config = {
"model_type": "transformer",
"num_layers": 3,
"hidden_size": 128,
}conv2d: 2D seperable depthwise conv net with lat/lons as the spatial dimensions
model_config = {
"model_type": "conv2d",
"num_layers": 7,
"hidden_size": 176,
"kernel_size": 3,
}To train:
main(**config.config)After training the model is saved under lightning_logs/version_XX .
All the parameters are also saved under
lightning_logs/version_XX/hparams.yaml
To use a model saved under saved under lightning_logs/version_XX
pass the checkpoint path to ckpt argument and all the configuration
will automatically load
config = Config(
{
"mode": "predict",
"dataset_params": {
"dataset": "toy",
"inputs": inputs,
"outputs": outputs,
"batch_size": 4096,
},
"trainer_params": {"gpus": [gpu], "max_epochs": 100},
"model_params": {
"ckpt": "lightning_logs/version_XX",
},
}
)
main(**config.config)Predictions file will be written out to the experiment checkpoint.
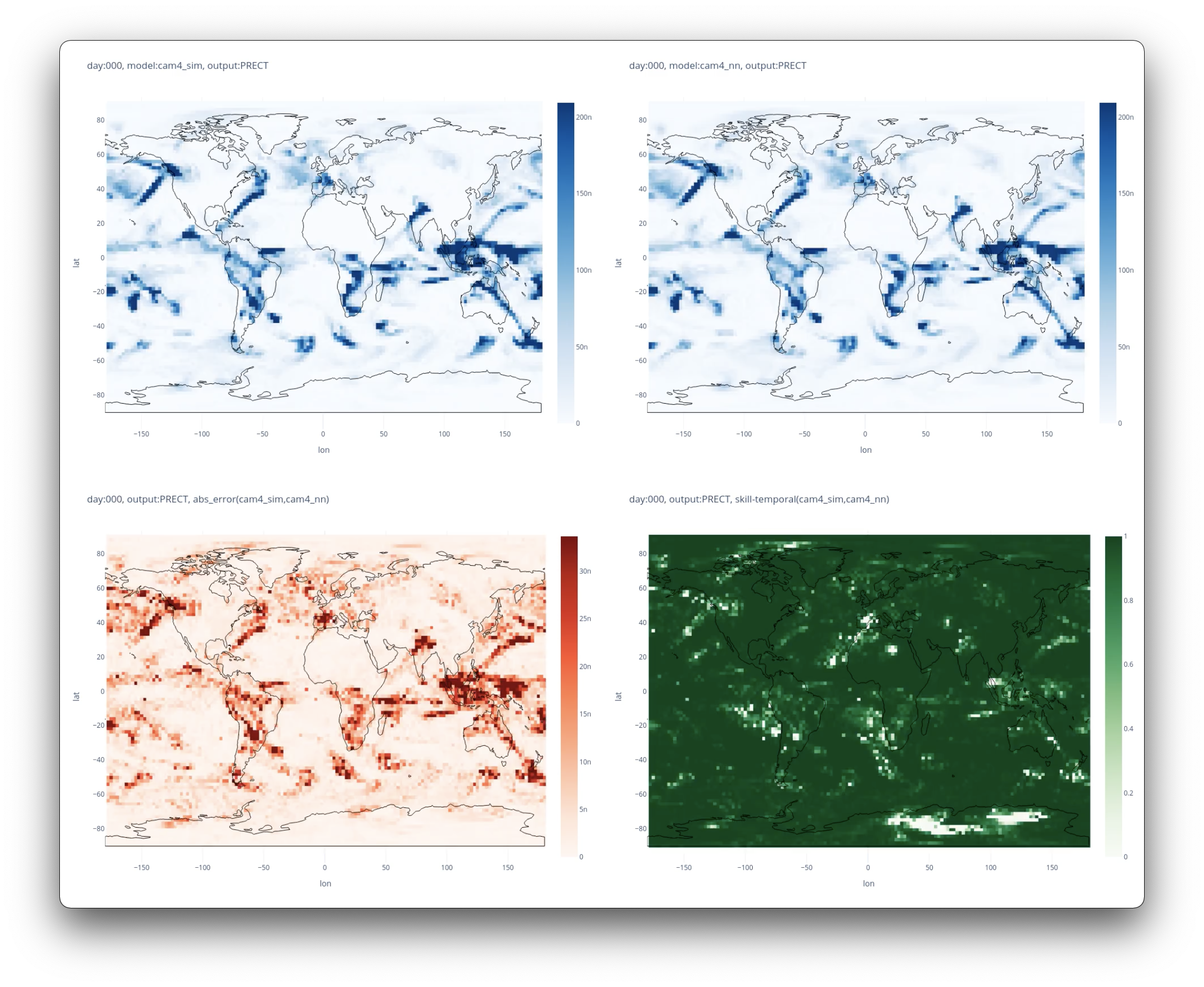
Plots will be saved in the experiment directory
from gaia.plot import save_diagnostic_plot, save_gradient_plots
save_gradient_plots(model_dir, device = f"cuda:{gpu}")
save_diagnostic_plot(model_dir)Export pretrained pytorch model to a torchscript checkpoint to be loaded into the intergrated hybrid model.
from gaia.export import export
model_dir = "lightning_logs/version_3"
export_name = "export_model_cam4.pt"
export(model_dir, export_name)To use a pretrained model:
config = Config(
{
"mode": "predict",
"dataset_params": {
"dataset": "toy",
"inputs": inputs,
"outputs": outputs,
"batch_size": 4096,
},
"trainer_params": {"gpus": [gpu], "max_epochs": 100},
"model_params": {
"ckpt": "path_to_checkpoint_directory",
},
}
)
main(**config.config)For lower level model access, you can load it directly:
from gaia.models import TrainingModel
model = TrainingModel.load_from_checkpoint(get_checkpoint_file(model_dir))Download pre-trained models: