Mapping out the coarse-grained connectivity structures of complex manifolds (Genome Biology, 2019).
PAGA is available within Scanpy through: tl.paga | pl.paga | pl.paga_path | pl.paga_compare.
Below you find links to all central example notebooks, which also allow reproducing all main figures of the paper. If you start working with PAGA, go through blood/paul15.
| notebook | system | details | reference | figure |
|---|---|---|---|---|
| blood/simulated | hematopoiesis | simulated | Krumsiek et al., Plos One (2011) | 2a |
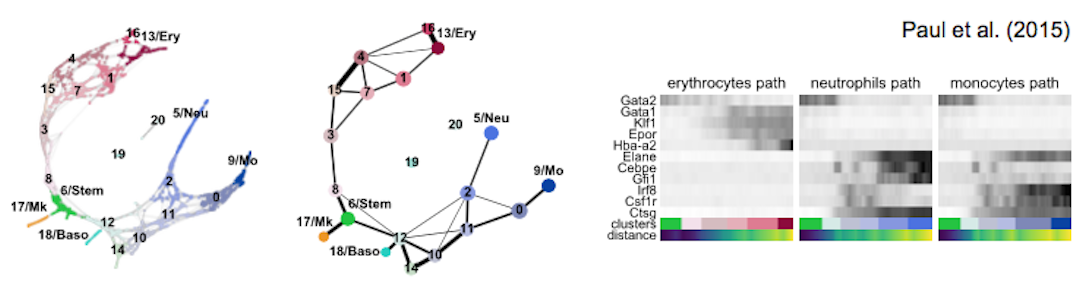
| blood/paul15 | murine hematopoiesis | 2,730 cells, MARS-seq | Paul et al., Cell (2015) | 2b |
| blood/nestorowa16 | murine hematopoiesis | 1,654 cells, Smart-seq2 | Nestorowa et al., Blood (2016) | 2c |
| blood/dahlin18 | murine hematopoiesis | 44,802 cells, 10x Genomics | Dahlin et al., Blood (2018) | 2d |
| planaria | planaria | 21,612 cells | Plass et al., Science (2018) | 3 |
| zebrafish | zebrafish embryo | 53,181 cells | Wagner et al., Science (2018) | 4 |
| 1M_neurons | neurons | 1.3 million cells, 10x Genomics | 10x Genomics (2017) | S12 |
| deep_learning | cycling Jurkat cells | 30,000 single-cell images | Eulenberg et al., Nat. Commun. (2017) | S14 |
All supplemental figures of the paper can be reproduced based on the following table.
| notebook | description | figure |
|---|---|---|
| connectivity_measure | connectivity measure | S1, S2, S3 |
| robustness | robustness and multi-resolution capacity | S4, S5 |
| comparisons/simulated_data | comparisons for simulated data | S6, S7 |
| comparisons/paul15_monocle2 | comparison Monocle 2 for Paul et al. (2015) | S8 |
| comparisons/nestorowa16_monocle2 | comparison Monocle 2 for Nestorowa et al. (2016) | S9 |
| embedding_quality | quantifying embedding quality | S10 |
| simulation | simulating hematopoiesis | S11 |
| 1M_neurons | neurons, 1.3 million cells, 10x Genomics, 10x Genomics (2017) | S12 |
| blood/paul15 | annotation of louvain clusters using PAGA | S13 |
| deep_learning | cycling Jurkat cells, 30,000 single-cell images, Eulenberg et al., Nat. Commun. (2017) | S14 |