-
Notifications
You must be signed in to change notification settings - Fork 36
knn4qa @ CIKM 2016
- Introduction * Overview * Pipeline architecture * NMSLIB: modus operandi overview * Bootstrapping & prerequisites * Data and software layout
- Data processing
- Experimentation
Here we describe how to reproduce results reported in our paper L. Boytsov, D. Novak, Y. Malkov, E. Nyberg (2016). Off the Beaten Path: Let’s Replace Term-Based Retrieval with k-NN Search, to appear in proceedings CIKM'16. The included code can be also used to partially reproduce results from the paper: M Surdeanu, M Ciaramita, H Zaragoza. Learning to rank answers to non-factoid questions from web collections Computational Linguistics, 2011.
We use three collections:
- L6 - Yahoo Answers Comprehensive version 1.0 (about 4.4M questions);
- L5 - Yahoo Answers Manner version 2.0 (about 142K questions), which is a subset of L6 created by Surdeanu al. 2011.
- Stack Overflow (about 8.8M answered questions).
The Yahoo Answers Manner collection, henceforth simply Manner, is too small to test retrieval algorithms. We use it only to ensure that our ranking model is state-of-the-art. To this end, we compare performance of this model against the previously published result by Surdeanu et al 2011. Only two larger collections: Yahoo Answers Comprehensive (henceforth Comprehensive) and Stack Overflow are used to compare generic k-NN search algorithms against classic term-based retrieval.
Yahoo Answers Comprehensive contains about 4.4M questions each of which has an answer. Furthermore, there is almost always a so-called best answer, which is selected by community members. In Stack Overflow, there are 8.8M answered questions, but the best answer is not always selected by an asker. Such questions are discarded leaving us with 6.2M questions.
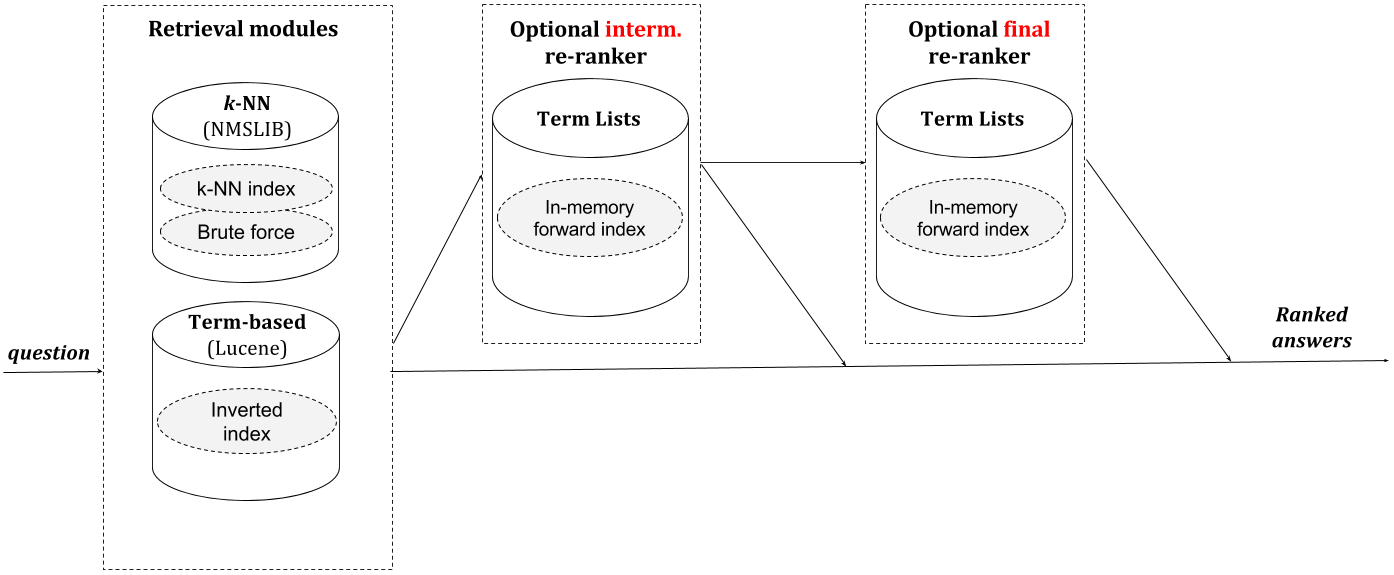 Our retrieval pipeline is reasonably efficient and works in a multi-threaded mode.
Pipeline's architecture is outlined in the above figure. Each pipeline has a single retrieval module and one or two re-ranking modules. There may be multiple retrieval modules that generate a list of candidates. Most importantly, we use Apache Lucene and a special branch of the Non-Metric Space LIBrary (NMSLIB). The pipeline is reasonably efficient and works in a multi-threaded mode.
Our retrieval pipeline is reasonably efficient and works in a multi-threaded mode.
Pipeline's architecture is outlined in the above figure. Each pipeline has a single retrieval module and one or two re-ranking modules. There may be multiple retrieval modules that generate a list of candidates. Most importantly, we use Apache Lucene and a special branch of the Non-Metric Space LIBrary (NMSLIB). The pipeline is reasonably efficient and works in a multi-threaded mode.
Likewise, there can be different re-ranking modules. Re-ranking modules differ in feature generation approaches as well as in employed learning-to-rank algorithms. Because we use RankLib models one can use only the learning-to-rank algorithms that are implemented in RankLib (BTW, RankLib is cool). Note that the final re-ranking model can be any RankLib model including a non-linear award-winning LambdaMART. The intermediate re-ranking model can be only linear.
The Non-Metric Space Library (NMSLIB) is a cross-platform similarity search library and a framework for evaluation of similarity search methods. NMSLIB has a concepts of index (a search method), a concept of a space, and a concept of an object. At the very minimum, a space provides an implementation to compute the distance between two points as well as functions for data serialization and de-serialization.
This basic space functionality is sufficient for distance based indexing methods, which treat data points as unstructured objects together with a black-box distance function. The indexing and searching processes of distance-based methods exploit only values of mutual object distances. There are no assumptions about distance values (e.g., non-negativity is not assumed) except that the closer are the points, the smaller is the distance. In particular, the nearest neighbor is a data set point with the smallest distance to the query.
NMSLIB can be extended by implementing new black-box "distance" functions and indexing methods.
The implementation process is outlined in the library manual.
In particular, for the purpose of our evaluation we have implemented a new configurable space codenamed QA1 (this space exists only in our special branch). This space implements the distance function, which mimics the linear similarity function produced by a Java re-ranker. To this end the space QA1 does the following:
- reads data (namely, a forward file) produced by a Java indexing application and can compute a variety of similarity functions;
- reads translation models produced by GIZA++ and linear machine learning models produced by RankLib.
- computes all major features (e.g., BM25 and IBM Model 1 scores) supported by a Java re-ranking module.
NMSLIB reads all data and models and builds an index optimized for k-NN search (alternatively it can rely on brute-force). The index can be optionally saved to disk and loaded from disk, if needed. NMSLIB then starts a multi-threaded TCP/IP server that answers queries submitted by our Java (retrieval) pipeline.
Important note: As mentioned previously, we use a slightly modified version of NMSLIB, which is kept as a separate branch). This branch differs in three ways:
- It contains the special space QA1 outlined above.
- It contains a slightly modified version of the search method
NAPPto work with the space QA1 most efficiently. - A modified class
Objectthat introduces a concept of a payload.
The last two modifications are necessary to support an on-line creation of a special inverted index for each query (as described in our paper).
Creating this index takes time, but it allows to greatly speed up computation of IBM Model 1. The inverted index is stored in the form of the Object
payload (that exists only in this special branch). In addition, we modify NAPP so that it creates these special inverted indices while
working with the space QA1.
One important caveat is that the time to create the inverted index is, of course, included into the overall query execution time in the main experiments. However, due to technical limitations it is not currently included when query time is measured using NMSLIB, i.e., in tuning experiments. For this reason, we have introduced a special header-file flag that disables/enables the use of payloads (useHashBasedPayloads). During tuning, we use a header where useHashBasedPayloads=0. This is not an ideal approach, yet, it still allows us to get a pretty good idea as to what set of parameters is good.
Most importantly, we need Oracle Java 8 and GCC version 3.x (x>=8) or 4.x. This project has a lot of other moving parts, which might be a bit overwhelming. Fortunately, these parts are open-source and are relatively to easy to install. To further simplify things, we have created a basic installation script scripts/bootstrap/install_packages_ubuntu14.sh for Unbuntu 14. This script installs nearly all the software and creates all the data directories. We have tested this script and (most other scripts) on a virtual Amazon host.
Before automatic installation, however, a few manual steps need to be done. First, you may need to install bash and wget (although they now seem to be included in a basic Ubuntu 14 installation). Second, you need to download the installation script itself, e.g.:
wget https://raw.githubusercontent.com/oaqa/knn4qa/cikm2016/scripts/bootstrap/install_packages_ubuntu14.sh -O install_packages_ubuntu14.sh
chmod +x install_packages_ubuntu14.sh
Note that you download the installation script from branch cikm2016!
Third, you need to install Oracle Java 8 and make sure that both java and javac are in the path (download Java SDK 8 from the Oracle download page). More specifically, you want the downloaded version of Java to have precedence over all other versions installed on your server (unless you use the right version of Java already). In addition, it is often helpful (for Maven), if you make an environment variable JAVA_HOME point to the installation directory of Java. For example, if you unpacked JDK 1.8 to your home directory, you will need to make the following modifications to environment variables (you can add these lines to your .bashrc file):
export PATH=jdk1.8.0_11/bin:$PATH
export JAVA_HOME=~/jdk1.8.0_11/
After these basic installation steps are completed, the main installation process can be started as follows:
bash install_packages_ubuntu14.sh soft data 8
Note that the first argument is the software installation directory; the second argument is the data installation directory; the third argument is the number of parallel processes to build C/C++ projects (however, we build Apache Thrift using a single process, because compilation process may fail otherwise). The script install_packages_ubuntu14 installs a number of software packages, most importantly, GNU C/C++, Maven, GIZA++, Python, Apache Thrift version 0.9.2, evaluation utility trec_eval version 9.0.4, Sparse Hash, and a patched version of ClearNLP 2.0.2. Importantly, the script uses sudo, so your user should have sudo privileges. You may also need to enter a password. This may be required for sudo. However, you will not need to enter anything if you use Amazon AWS.
To verify the correctness of install as well as to demonstrate how to run a full set of experiments (except tuning), we implemented a small-scale end-to-end testing script. Please, see the respective section for more detail. This script consumes minimally processed data. The process of producing this data is described in the previous subsection.
We have two major pieces of software that interact with each other: the re-ranking modules written in Java and the k-NN retrieval modules written in C++. The Java code comes from this repository. The k-NN retrieval modules comes from a special branch of NMSLIB. There are also two major locations on disk: a software installation directory and a data directory.
The software installation directory keeps our Java and C++ code, which are in subdirectories knn4qa and nmslib respectively.
It is also the directory where we install GIZA++. A bunch of other software modules is installed to standard locations.
The subdirectory knn4qa (in the main installation directory) is a main directory. When not stated otherwise, it is a starting point for all processing scripts.
The data directory structure created by the bootstrapping script looks as follows (see below for a description):
├── input
│ ├── compr
│ ├── ComprMinusManner
│ ├── manner
│ └── stackoverflow
├── output
│ ├── compr
│ │ ├── dev1
│ │ ├── dev2
│ │ ├── test
│ │ ├── train
│ │ └── tran
│ ├── ComprMinusManner
│ │ └── tran
│ ├── manner
│ │ ├── dev1
│ │ ├── dev2
│ │ ├── test
│ │ └── train
│ └── stackoverflow
│ ├── dev1
│ ├── dev2
│ ├── test
│ ├── train
│ └── tran
├── lucene_index
│ ├── compr
│ ├── ComprMinusManner
│ ├── manner
│ └── stackoverflow
├── memfwdindex
│ ├── compr
│ ├── ComprMinusManner
│ ├── manner
│ └── stackoverflow
├── tran
│ ├── compr
│ ├── ComprMinusManner
│ ├── manner
│ └── stackoverflow
├── WordEmbeddings
│ ├── Complete
│ ├── compr
│ ├── ComprMinusManner
│ ├── manner
│ └── stackoverflow
└── nmslib
├── compr
│ ├── headers -> /home/ubuntu/soft/knn4qa/scripts/nmslib/meta/compr/headers/
│ ├── memfwdindex -> /home/ubuntu/data/memfwdindex/compr
│ ├── models -> /home/ubuntu/soft/knn4qa/scripts/nmslib/meta/compr/models/
│ ├── index
│ ├── pivots
│ ├── queries
│ ├── tran -> /home/ubuntu/data/tran/compr
│ └── WordEmbeddings -> /home/ubuntu/data/WordEmbeddings/compr
└── stackoverflow
├── headers -> /home/ubuntu/soft/knn4qa/scripts/nmslib/meta/stackoverflow/headers/
├── memfwdindex -> /home/ubuntu/data/memfwdindex/stackoverflow
├── models -> /home/ubuntu/soft/knn4qa/scripts/nmslib/meta/stackoverflow/models/
├── index
├── pivots
├── queries
├── tran -> /home/ubuntu/data/tran/stackoverflow
└── WordEmbeddings -> /home/ubuntu/data/WordEmbeddings/stackoverflow
The input directory contains several sub-directories (one for each collection).
Input files of the same collections are randomly split into several subsets (including training, development, and testing subsets). The directory output contains the output of the annotation process. Similarly to the structure of input directory, there is a sub-directory for each collection. For more details, see the section Data Processing below.
The contents of output directory are used to produce two types of indices: a Lucene index and a forward file, which are stored in directories lucene_index and ``memfwdindex'', respectively. Again, there is a separate sub-directory for each collection. For details of the indexing processing please refer to the respective section.
The directory tran contains translation models (IBM Model 1). This topic is covered in more detail in the respective section.
The directory WordEmbeddings contains collection-specific word embeddings. To produce these embeddings, you first need to download word2vec embeddings produced from Google News corpus. Download the file GoogleNews-vectors-negative300.bin and place it into directory WordEmbeddings/Complete (unfortunately, this cannot be done automatically, b/c the model is stored in Google drive). Afterwards, one should build collection-specific word embeddings as it is described in this section.
The directory nmslib is used by k-NN search retrieval modules implemented in NMSLIB. For every large collection, there is sub-directory, where we gather related data pieces, which are mostly linked to existing data directories. Consider, e.g., the directory structure for Stack Overflow:
stackoverflow
├── headers -> /home/ubuntu/soft/knn4qa/scripts/nmslib/meta/stackoverflow/headers/
├── memfwdindex -> /home/ubuntu/data/memfwdindex/stackoverflow
├── models -> /home/ubuntu/soft/knn4qa/scripts/nmslib/meta/stackoverflow/models/
├── index
├── pivots
├── queries
├── tran -> /home/ubuntu/data/tran/stackoverflow
└── WordEmbeddings -> /home/ubuntu/data/WordEmbeddings/stackoverflow
- Directory
headersis a link to the repository directory that contains the so-called collection headers. A header is an input meta-file used by NMSLIB to understand how to load and use the data set. In addition to specifying the location of data file, a header describes a similarity model. If necessary, it should also specify the location of word embeddings and translation files. Only three header files are essential:header_bm25_text,header_exper1, andheader_avg_embed_word2vec_text_unlemm. The first two headers are used by the search method NAPP (see our paper for details). The fileheader_bm25_textinstructs NMSLIB to use the similarity model BM25. The fileheader_exper1instructs NMSLIB to use the similarity model IBM Model1+BM25 (a weighted combination of two similarity scores). The fileheader_avg_embed_word2vec_text_unlemminstructs NMSLIB to use the cosine similarity between averaged word embeddings. In this case, we use an indexing/search algorithm SW-graph. - The second directory is a link to the directory containing a forward index. NMSLIB reads a forward index and constructs in-memory indices optimized for fast k-NN retrieval. Alternatively, NMSLIB can answer queries in the brute-force mode, which does not require creation of an index.
- Directory
modelsis a link to a repository directory that contains an already pre-trained linear model in the RankLib format. NMSLIB can use this model to compute the non-metric similarity/distance function. Note that NMSLIB can use only linear RankLib models produced by the coordinate ascent algorithm. - Directory
indexcontains k-NN indices created by NMSLIB. More exactly, these indices are stored in additional sub-directories, whose names depend on the type of the similarity method and its parameters. - Directory
pivotscontains generated pivots. Using such pivots allows us to greatly speed up the k-NN search method NAPP. Pivots are created by the scriptscripts/nmslib/gen_nmslib_pivots.sh. - Directory
queriescontains queries (e.g., fromdev1subset) in the format that NMSLIB can understand. These queries are used for tuning only and are not required to reproduce most of the results. - Directory
transimply links to the directory with the translation model (IBM Model 1) for Stack Overflow. If needed, NMSLIB will read the translation model and use it compute the distance/similarity. - Directory
WordEmbeddingsis a link to a collection-specific directory that contains collection-specific word embeddings (see the section Building derivative data for more details). The content of this directory is used by NMSLIB to compute averaged word embeddings as well as the cosine similarity between averaged word embeddings.
Yahoo Answers collections are available through Yahoo! WebScope and can be requested by researchers from accredited universities only. Unfortunately, Yahoo also forbids publishing any derivative data for this data. Manner comes in one file. Comprehensive comes in two files, which need to be concatenated into a single file (refer to Yahoo Answers README for details).
The Stack Overflow collection is freely available for download.
We use the dump dated March 10 2016 from which we remove all the code (the content marked by the tag code).
Because we started working with Yahoo Answers collection first and added Stack Overflow later, our system understands data only in Yahoo Answers format.
When faced with the necessity to process Stack Overflow as well, we proceeded exactly as a mathematician in the joke: We reduced the problem to the already solved one by implementing a converter from Stack Overflow to Yahoo Answers format.
How does a mathematician or a physicist prepare tea?
Both of them pour water into the pot, light the flame, boil the water, infuse the leaves.
What is the difference in the solution, if the pot is already filled with water?
The physicist lights the flame, boils the water, infuses the leaves.
The mathematician pours the water out, thereby reducing the problem to the previous, already solved one.
The converter is invoked by the script scripts/data/run_convert_stackoverflow.sh.
For convinence, we provide the already created version in the Yahoo Answers format.
Should you decide to run conversion yourself, do not forget to specify the option -exclude_code.
Each collection is randomly split into several subsets, which include training, two development (dev1 and dev2), and testing subsets.
In the case of Comprehensive and Stack Overflow, there is an additional subset (denoted tran) that is used to learn IBM Model 1.
The answers from tran are indexed, but the questions are discarded after learning Model 1 (i.e, they are not used for training and testing).
In the case of Manner, IBM Model 1 is trained on a subset of Comprehensive from which we exclude QA pairs that belong to Manner (using the script scripts/data/diff_collect.sh).
The split of Manner mimics the setup Surdeanu et al. 2011. Assuming that raw data is stored in directory raw and the split collection stored in the directory input/manner using file names prefixed with manner, the splitting script can be invoked as follows (note that parameter -p specifies comma-separated probabilities and parameter -n names of respective collections):
scripts/data/split_input.sh -i raw/manner-v2.0/manner.xml.bz2 -o input/manner/manner-v2.0 -p 0.60,0.05,0.15,0.20 -n train,dev1,dev2,test
This commands would create the following files corresponding to the training, development, and testing subsets:
manner-v2.0_train.gz
manner-v2.0_dev1.gz
manner-v2.0_dev2.gz
manner-v2.0_test.gz
For Comprehensive and Stack Overflow we use the following splitting commands:
scripts/data/split_input.sh -i raw/FullOct2007.xml.bz2 -o input/manner/comprehensive-Oct2007 -p 0.048,0.0024,0.0094,0.0117,0.9285 -n train,dev1,dev2,test,tran
scripts/data/split_input.sh -i raw/PostsNoCode2016-04-28.xml.bz2 -o input/stackoverflow/StackOverflow -p 0.048,0.0024,0.0094,0.0117,0.9285 -n train,dev1,dev2,test,tran
Important notes::
- Please, pay attention to the prefixes of output files**!
The collection processing scripts (see below) assume that input files are prefixed using this name and are located in the directory
input/<collection mnemonic name>. - As you may notice the above splits create more than 10K test queries for Comprehensive and Stack Overflow, while we use 10K in paper. For this reason, either has to use a different collection split or specify the maximum number of queries for testing scripts. We explain this below.
In the case of StackOverflow, we provide the already split collection in Yahoo Answers format ourselves. We used this split collection ourselves: train,dev1, dev2, test, tran.
Preprocessing of data is done by an annotating Java pipeline that relies on Apache UIMA. As a reminder, annotations are a commonly used formalism in NLP. We create several linguistically motivated textual representations of questions and answers [103], which we call fields. A field may include word tokens, syntactic dependencies, ProbBank style semantic role labels for verbal predicates (SRLs), or WordNet super-senses (like it is done by Surdeanu et 2011).
An example of the complete annotation (i.e., all fields included is given in this table):
| Type | Field name | Sample question |
|---|---|---|
| Original text | How does the President nominate Supreme court justices? | |
| Lemmatized text | text | president nominate supreme court justice |
| Unlemmatized text | text_unlemm | does president nominate supreme court justices |
| Bigrams (lemmatized) | bigram | president nominate nominate supreme supreme court court justice |
| Semantic roles (unlabeled) | srl | nominate president nominate justice |
| Semantic roles (labeled) | srl_lab | nominate A0 president nominate A1 justice |
| Dependencies (unlabeled) | dep | nominate president court supreme justice court nominate justice |
| WordNet super-senses | wnss | verb.social noun.person verb.communication noun.group noun.attribute |
However, by default a pipeline creates only three non-empty fields: text, text_unlemm, and bigram.
In other words, it annotates text by marking sentences, invididual tokens, obtains token lemmas, and concatenates adjacent lemmas into bigrams.
Additional annotations did not prove to be useful, though.
For this reason, we do not enable them by default. However, the pipeline can also be configured (see a description below) to create all possible fields.
For each field and collection subset, the pipeline creates two separate output file: one for answers and another for questions.
For example, we have files answer_text (for token lemmas), answer_text_unlemm (for original tokens), answer_bigram (lemmatized bigrams), answer_dep (for dependency parse), question_text, question_text_unlemm, and so on.
Three additional files are also created: SolrQuestionFile.txt, SolrAnswerFile.txt, and ``qrels.txt''.
The first two files contain the combination of all fields for questions and answers respectively.
These files are fed to querying and indexing applications.
The last file contains relevance judgements.
As a reminder, the best answer is the only relevant document for a given question.
Before starting the annotation process, we first create output directories using the script scripts/data/create_output_dirs.sh. It creates a number of sub-directories inside the data directory output:
output/
├── compr
│ ├── dev1
│ ├── dev2
│ ├── test
│ ├── train
│ └── tran
├── ComprMinusManner
│ └── tran
├── manner
│ ├── dev1
│ ├── dev2
│ ├── test
│ └── train
└── stackoverflow
├── dev1
├── dev2
├── test
├── train
└── tran
The annotating script assumes the following:
- These output directory exists.
- The input is stored in the directory
input/manner(or more generally ininput/<collection mnemonic name>). Additionally, we assume that input data files have above-mentioned prefixes (manner-v2.0,comprehensive-Oct2007,StackOverflow) and suffixes (_train,_dev1,_dev2,_test) indicating the type of collection subsets. If all the naming conventions are fulfilled (for the collection that contains Comprehensive questions not present in Manner the file should be namedComprMinusManner.gz), the annotating script can be invoked as follows:
scripts/uima/run_annot_pipeline.sh manner
scripts/uima/run_annot_pipeline.sh compr
scripts/uima/run_annot_pipeline.sh ComprMinusManner
scripts/uima/run_annot_pipeline.sh stackoverflow
When finished successfully, the script prints execution statistics, which includes something like:
Component Name: aeWrapperAggreg
Event Type: End of Batch
Duration: 20ms (0.03%)
Annotation seem to have finished successfully!
In the end, you should see the message Annotation seem to have finished successfully!.
Notes on configuration:
- The annotating script is multi-threaded and uses 4 threads by default. The number of threads can be increased by modifying all (or collection-specific) UIMA configuration files
collection_processing_engines/cpeAnnot*. To this end, you need to find the entry:<casProcessors casPoolSize="4" processingUnitThreadCount="4">and replace 4 with a desired number of threads in BOTH attributes. Note that this would also increase memory consumption! - As mentioned above, it is possible to invoke a fuller annotation process. To this end, one needs to modify all (or collection-specific) UIMA configuration files
analysis_engines/aeWrapperAggreg*. To this end, you need to find the following entry:<import name="descriptors.analysis_engines.aeAnnotAggregSimple"/>and replace it with<import name="descriptors.analysis_engines.aeAnnotAggreg"/>. Note that a fuller annotation process requires installing WordNet and a fixed version of ClearNLP (see a description in prerequisites section).
First use a helper script scripts/index/create_index_dirs.sh to create index directories for all collections. There are two types of indices that need to be created for every collection: a Lucene index and a forward index. The forward file will be loaded by querying applications (NMSLIB and/or Java pipeline) into memory. Here are the sample commands for Comprehensive:
scripts/index/create_lucene_index.sh compr
scripts/index/create_inmemfwd_index.sh compr
Please, disregard numerous messages like:
Warning: empty field 'WNSS_bm25' for document '5409-4'
Translation (IBM Model 1) models are built using GIZA++, which is downloaded and compiled by the installation script. This script also creates an output directory tran. IBM Model 1 can be created for any field. However, not all field models are equally effective. We find that it is best to create the model for lemmatized text. The model for unlemmatized text can be sometimes a bit better. Yet, it also consumes more space and requires more memory to build. Building the model requires specifying the number of iterations. Like Surdeanu et al 2011, we use only five iterations. We also observe that adding more iterations does not drastically reduce training perplexity (i.e., the model seem to converge quickly).
Here are three sample commands to build translation models for collections Manner, Compr, and Stack Overflow, respectively:
scripts/giza/create_tran.sh output tran ComprMinusManner tran text /home/ubuntu/soft/giza-pp 0 5
scripts/giza/create_tran.sh output tran ComprMinusManner tran text_unlemm /home/ubuntu/soft/giza-pp 0 5
scripts/giza/create_tran.sh output tran compr tran text /home/ubuntu/soft/giza-pp 0 5
scripts/giza/create_tran.sh output tran compr tran text_unlemm /home/ubuntu/soft/giza-pp 0 5
scripts/giza/create_tran.sh output tran stackoverflow tran text /home/ubuntu/soft/giza-pp 0 5
scripts/giza/create_tran.sh output tran stackoverflow tran text_unlemm /home/ubuntu/soft/giza-pp 0 5
Note several important things:
- Building the model requires data files produced by annotation pipelines. These data files are stored in the subdirectories of the directory
outputas it is described above. For example, the files belonging to the collection Comprehensive are stored inoutput/compr. - All scripts using translation models assume that the number of iterations is 5. Don't change this value! Furthermore, there seems to be little benefit of using more iterations, in particular, because training perplexity does not decrease much after iteration 3-4.
- The so-called translation files representing IBM Model 1 are stored in subdirectories of
tran. For example, in the case of Stack Overflow and fieldtextthey are stored intran/stackoverflow/text.orig. The directorytext.origcontains unpruned model. However, this model is too large to be practical. Thus, we prune it by removing entries with small translation probabilities. In the next section, we explain how pruning is carried out. - We build the model for two fields:
text, andtext_unlemm. The latter model is currently used only to retrofit word embeddings (see below).
The two larger collections: Comprehensive and Stack Overflow are split into several parts and the largest part (namely, tran) is used to build translation models.
However, the translation model for collection Manner is built from ComprMinusManner, which is the collection that contains Comprehensive question and answers not present in Manner.
Finally, note that in our example, we run the tests on Amazon AWS and use /home/ubuntu/soft/ as the installation directory.
Hence, the location of GIZA++ is /home/ubuntu/soft/giza-pp.
This may need to be replaced with a path to GIZA++ on your computer/server.
Because translation tables are large, we have to prune them. This can be done only after the forward index is created.
To simplify things, generation of pruned translation tables is a part of the script that generates derivative data (see the next section).
This script calls scripts/giza/run_filter_tran_table.sh, which, in turn, invokes a pruning Java application.
The application has several parameters.
The most important are: the minimum translation probability and the number of most frequent words to be included.
For convenience, in the case of Stack Overflow only, we publish already pruned translation tables for the fields text and text_unlemm. Read our words of caution: If you ever use already precomputed translation tables, you have to use the same split of the corpus as we did. Otherwise, some of the QA pairs that we used to train IBM Model 1 will end up in your training, testing, or development sets (which would amount to using part of your training data for testing)!
The derivative data includes the pruned translation tables and retrofitted word embeddings. The embeddings are retrofitted using IBM Model 1 as a relational lexicon. In other words, we take a set of pre-trained embeddings and try to modify them (while still preserving original values to a good degree) so that embeddings become closer for pairs of words that have high translation probabilities.
We use word2vec embeddings trained on Google News corpus.
Download the file GoogleNews-vectors-negative300.bin and place it into directory WordEmbeddings/Complete (if there are two words with different casing, we keep only a lowercase version).
Then, you can generate derivative data (for three collections) as follows:
scripts/data/gen_derivative_data.sh manner
scripts/data/gen_derivative_data.sh compr
scripts/data/gen_derivative_data.sh stackoverflow
Important note: The script expects that translation tables are created for the fields text and text_unlemm only.
If you want to create additional translation tables, e.g., for bigram, you would need to modify the script by adding this field to the variable FIELD_LIST, e.g.:
FIELD_LIST="text text_unlemm bigram"
The pruning software takes translation tables from the subdirectories text.orig and text_unlemm.orig.
Then, it places pruned version into the subdirectories text and text_unlemm respectively.
Using generated pivots allows us to greatly speed up the k-NN search method NAPP.
Pivots are created by the script scripts/nmslib/gen_nmslib_pivots.sh separately for each collection:
scripts/nmslib/gen_nmslib_pivots.sh /home/ubuntu/data compr
scripts/nmslib/gen_nmslib_pivots.sh /home/ubuntu/data stackoverflow
Important note: You have to create a forward index before generating pivots. Just in case, you can download pivots generated for Stack Overflow here.
Queries are the same queries as those used by our retrieval pipeline.
The difference is that scripts/nmslib/gen_nmslib_queries.sh produces queries in the NMSLIB format.
These queries are used for tuning only (see the respective section) and are not required to reproduce most of the results.
Queries are created using the script scripts/nmslib/gen_nmslib_queries.sh separately for each collection and each subset.
Because we use only dev1 for tuning, all the necessary queries are generated as follows:
scripts/nmslib/gen_nmslib_queries.sh /home/ubuntu/data/ compr dev1
scripts/nmslib/gen_nmslib_queries.sh /home/ubuntu/data/ stackoverflow dev1
Before running experiments make sure you generated the following data and indices:
- Pruned translation models.
- Collection-specific word embeddings.
- Lucene index.
- Forward index.
For an example of executing all these steps, please, refer to the script scripts/bootstrapping/small_scale_test.sh.
To verify the correctness of install as well as to demonstrate how to run a full set of experiments (except tuning), we implemented a small-scale end-to-end testing script scripts/bootstrapping/small_scale_test.sh.
This script randomly re-splits the collection, extracts, annotates small subsets of original data, and runs a few auxilliary scripts that process data. After that, scripts/bootstrapping/small_scale_test.sh carries out all major experiments.
Because experiments are run at small scale, they take relatively short time to execute. Perhaps, the longest step is splitting of the collection. It takes long because the splitting procedure operates on complete collections. We tested scripts/bootstrapping/small_scale_test.sh on virtual Amazon AWS host (instance r3.large). It took nearly 13 hours to finish.
The script scripts/bootstrapping/small_scale_test.sh has three arguments, which include following:
- the software installation directory
- the data directory 3 the directory containing minimally processed data. The process of producing this data is described in the previous subsection.
Note the following things:
- The script
scripts/bootstrapping/small_scale_test.shreplaces some of the UIMA descriptors with descriptors that annotate only a subset of data. We do our best to restore original descriptors when the process terminates. However, such restoration cannot be guaranteed. Make sure to double check that the content ofsrc/main/resources/descriptorshas not changed usinggit diff. - The script
scripts/bootstrapping/small_scale_test.shexpects specific names of inputrawfiles:"manner.xml.bz2for Manner,FullOct2007.xml.bz2for Comprehensive, andPostsNoCode2016-04-28.xml.bz2for Stack Overflow.
Our retrieval pipeline can use multiple feature generators, which can be combined with learning-to-rank algorithms provied by RankLib. The employed set of features is taken largely from the paper of Surdeanu et al. 2011. Basically, features include TF*IDF and Model 1 scores for various fields. However, because our implementation of these features is diffferent, we carry out our own effectiveness evaluation on Manner and Comprehensive (using respective on development sets). The effectiveness of the features and learned models can be tested using the Java pipeline only
There are a number of low-level training and testing scripts, which we do not cover in detail. Using these scripts requires a rather detailed understanding of our code. Yet, we want to highlight two high-level scripts that cover two basic scenarios:
- Building a linear model for a given set of features and verifying its effectiveness
- Testing effectiveness of several pre-trained models
Both scripts first use Lucene to retrieve candidate records. Then, an in-memory forward index is used to generate features for these candidate records. These features can be saved to a file to be further used to train a learning-to-rank models. The features can also be used by a re-ranker (relying on an already existing learning-to-rank model) to re-rank candidate entries.
The first scenario is covered by the script scripts/exper/run_feature_experiments.sh, which accepts three parameters.
The first parameter is the mnemonic name of the collection (manner, compr, or stackoverflow);
the second parameter is a feature description file;
and the third parameter specifies the number of experiments to be executed in parallel.
Here is an example of calling this script for Manner:
scripts/exper/run_feature_experiments.sh manner scripts/exper/feature_desc/bm25_dev1.txt 1
The feature description files has three space-separated fields, e.g.:
exper@bm25=text,text_unlemm,bigram+model1=text @ dev2
The last field specifies the name of the collection subset.
For feature experiments, it is always either dev1 or dev2.
The first field specifies the name of the feature extractor.
For feature experiments, we use a special configurable extractor whose name always starts with the prefix exper@.
Then, goes a list of generated features in a human-readable format.
For example, the string bm25=text,text_unlemm,bigram+model1=text instructs our software to use BM25 scores for fields text, text_unlemm, and bigram as well as Model 1 scores for the field text.
Different feature types are separated by plus-signs.
For one type of feature, we start the description with feature name (e.g., bm25 or model1) followed by the equation sign, which is further followed by a comma-separated list of fields.
The second field specifies the file name containing word embeddings.
The character '@' means that no embeddings are necessary.
Instead, one can specify, e.g., word2vec_retro_unweighted_minProb=0.001.txt.
For more examples of feature types, please, see feature description files in the subdirectories of scripts/exper/feature_desc.
Because the testing script may spawn several parallel processes, their output is not logged onto the screen. Where to find log files and experimental results can be inferred from the script output such as:
The number of CPU cores: 24
The number of || experiments: 1
The number of threads: 24
Max # of queries to use:
Number of parallel experiments: 1
Number of threads in feature extractors/query applications: 24
Started a process 7054, working dir: results/feature_exper//manner/dev1/none/@
Waiting for 1 child processes
Process with pid=7054 finished successfully.
Started a process 7173, working dir: results/feature_exper//manner/dev1/exper@bm25=text/@
Waiting for 1 child processes
Process with pid=7173 finished successfully.
Waiting for 0 child processes
============================================
2 experiments executed
0 experiments failed
============================================
The second scenario is covered by the script scripts/exper/test_final_lucene_simple.sh, which has a somewhat similar functionality. The main difference is that it uses a hardcoded set of precomputed models.
The results are placed into sub-directories of directory results/final.
An exmple of running this script:
scripts/exper/test_final_lucene_simple.sh compr 10000
Note that the second argument is the maximum number of queries to execute. We limit this value to 10000 for Comprehensive and Stack Overflow, but not for Manner!
Re-runing tuning is not required to reproduce results.
Yet, we briefly describe the process.
The objective of tuning is to find optimal parameters that allows us to achieve maximum efficiency for a given level of k-NN recall.
Note that the tuning step relies only on intrinsic evaluation and does not use any extrinisic, i.e., IR metrics.
For this reason, we use a different starting point.
Specifically, we start the tuning script from inside the directory nmslib/similarity_search, e.g., as follows:
../../knn4qa/scripts/nmslib/tune_napp_bm25_text.sh compr 0 300 "13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29"
and
../../knn4qa/scripts/nmslib/tune_napp_exper1.sh compr 0 300 "13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29"
The first argument of the script specifies the mnemonic collection name; the second argument should be zero;
the third argument specifies the number of pivots to index (parameter numPivotIndex of NAPP),
and the last argument specifies the set of numPivotSearch arguments.
These are query-time parameters instructing NMSLIB regarding the minimum overlap between query and data points (in terms of the number of close pivots).
Note that for every unique value of numPivotIndex, the tuning script will create (a rather large) index in a sub-directory of indice/compr. The results will be in one of the sub-directories of results/tuning.
Note that this not required unless you modify header files and/or the code itself.
However, if you do so, you can use a debugging application invoked by the script scripts/query/run_debug_nmslib.sh.
The objective is to verify that similarity scores produced by the implementation of the similarity in NMSLIB are nearly the same as similarity scores produced by the Java pipeline (they cannot be exactly the same for a variety of good reasons).
This is not a trivial exercise, which requires a good understanding of our code.
In particular, this requires staring the NMSLIB query server manually. However, instead of specifying any fancy indexing method for k-NN search, one should start the server in brute-force search mode.
Yet, we want to inform you that such a debugging possibility exists, just in case.
Assume that you start the server on the same (local host) using port 9999. Then, you can run the debugging application as follows:
scripts/query/run_debug_nmslib.sh -cand_qty 200 -n 50 -max_num_query 1000 \
-embed_dir WordEmbeddings/compr \
-extr_type_interm exper@bm25=text,text_unlemm,bigram+model1=text+simple_tran=text \
-model_interm nmslib/compr/models/out_compr_train_exper@bm25=text,text_unlemm,bigram+model1=text+simple_tran=text_15.model \
-nmslib_addr localhost:9999 -nmslib_fields text,text_unlemm,bigram \
-q output/compr/dev1/SolrQuestionFile.txt \
-u lucene_index/compr \
-memindex_dir memfwdindex/compr -giza_root_dir tran/compr -giza_iter_qty 5
As described in our paper, the main objective is to measure efficiency-effectiveness trade-offs of k-NN search for three similarity models: BM25, BM25+Model 1, and the cosine similarity between averaged word embeddings. To do so, we evaluate performance of indexing approaches (SW-graph and NAPP) and compare it against performance of the brute-force search. As a reference point, we also compute effectiveness of the cosine similarity between TF*IDF vectors using the brute-force search.
There are several helpers scripts that facilitate running experiments each of which does the following:
- Starts the query server (you can check the log file
server.logif the server does not start); - Loads data and creates an index if necessary (indexing parameters are hardcoded, we select them based on results of tuning);
- Sets query-time parameters if necessary (indexing parameters are hardcoded, we select them based on results of tuning);
- Executes queries and evaluate effectiveness.
Each script accepts or one or two parameters.
Collection-specific scripts accept only the name of the collection subset (which is test in our experiments).
Other scripts have an additional parameter that specifies the collection mnemonic name.
In the following table we list all the scripts and indidcate which parameters they have.
In addition, to these parameters one can specify an optional parameter -max_num_query to limit the number of queries executed. In our paper, we ran only 10K queries for Comprehensive and Stack Overlow.
| Script name | Similarity model | k-NN search method | Positional parameters |
|---|---|---|---|
| test_nmslib_server_bruteforce_bm25_text.sh | BM25 | brute force | <collection> <test part> |
| test_nmslib_server_bruteforce_cosine_text.sh | TF*IDF cosine | brute force | <collection> <test part> |
| test_nmslib_server_bruteforce_exper1.sh | BM25+Model1 | brute force | <collection> <test part> |
| test_nmslib_server_bruteforce_swgraph.sh | Cosine embed. | brute force | <collection> <test part> |
| test_nmslib_server_napp_bm25_text_compr.sh | BM25 | NAPP | <test part> |
| test_nmslib_server_napp_bm25_text_stackoverflow.sh | BM25 | NAPP | <test part> |
| test_nmslib_server_napp_exper1_compr.sh | BM25+Model1 | NAPP | <test part> |
| test_nmslib_server_napp_exper1_stackoverflow.sh | BM25+Model1 | NAPP | <test part> |
| test_nmslib_server_swgraph.sh | Cosine embed. | SW-graph | <collection> <test part> |
Important note: There might be a situation (e.g., when you manually stop the script), when an NMSLIB query server continues to run in the background. In this case, when you run an experimental script again, it would complain about existing instance of the NMSLIB query server already running and stop. To fix this, you need to terminate the NMSLIB query server, e.g., as follows:
killall query_server