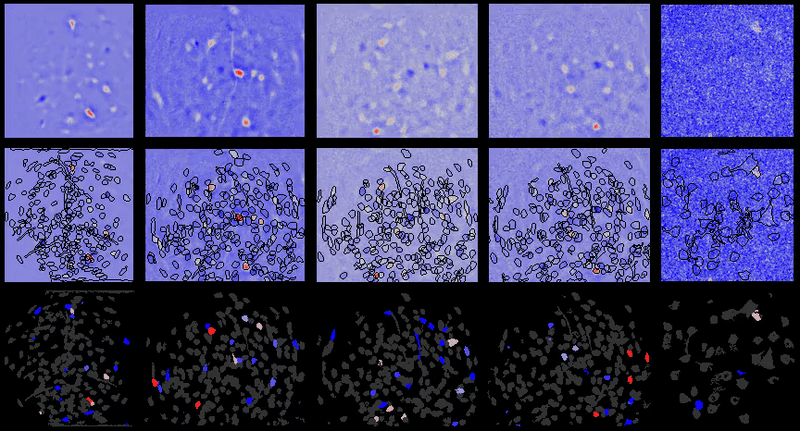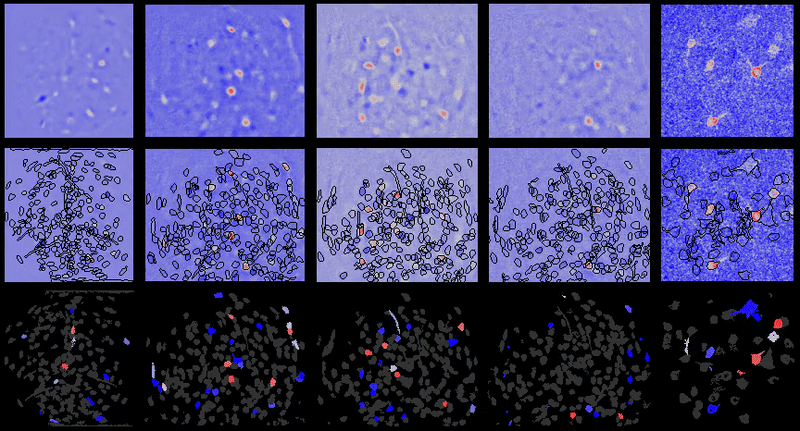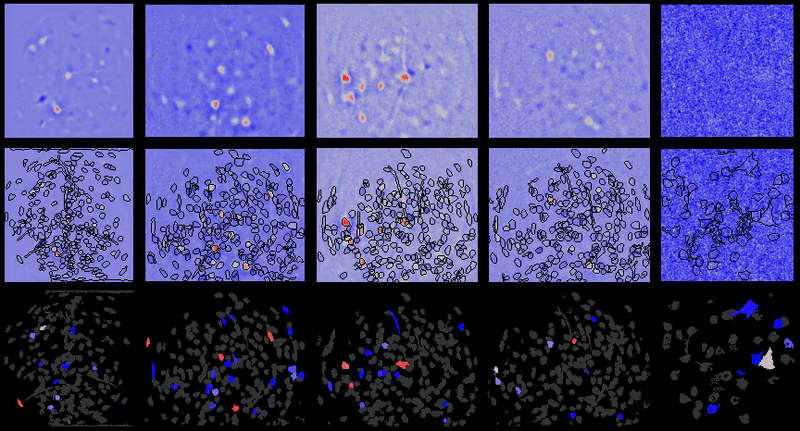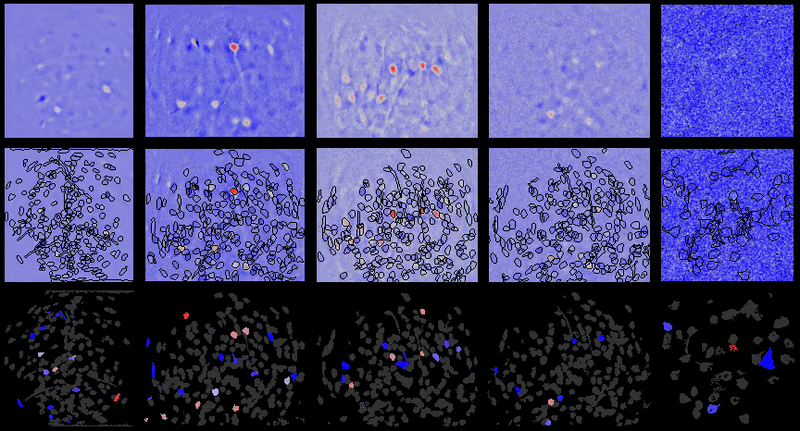
A table containing imaging analysis tools for biology and neuroscience, with a focus on calcium imaging.
Created by Biafra Ahanonu, PhD (HHMI Hanna Gray Fellow, Basbaum Lab, UCSF).
Calcium imaging analysis with CIAtah (https://github.com/bahanonu/ciatah).
The table can also be found at:
- https://bahanonu.com/brain/imaging_tools/
- https://bahanonu.com/syscarut/articles/233/
- If you would like to cite this table, see
Cite this repositoryin the rightAboutsection or https://zenodo.org/record/8349533.
Notes:
- I use cell extraction to refer to algorithms that perform cell segmentation and extract neural activity traces.
- In cases where the publication did not explicitly give the algorithm a name, made one based on the underlying method used.
- This table includes algorithms that simultaneously extract cell images/contours and reconstruct cell activity traces along with ones mainly focused on determining one or the other.
- Several calcium imaging related packages have also been included along with algorithms dealing with post-hoc handling of data or cell activity traces.
- Future versions of the repository will include table file (e.g. CSV) and basic LaTeX code so others can import or modify the table more easily going forward.
- Depending on monitor size and browser, scroll horizontally to see right-most table columns (e.g. websites/URLs).
- Any additional papers or algorithms that should be added or suggested updates to the table, leave a comment on the associated blog post or open an issue on the GitHub page, I want to make sure everyone’s brilliant work is acknowledged!
| # | Method | Year | Analysis pipeline | Notes/Code | Citation |
| 1 | PhaseCorrelation | 1996 | Motion correction. | • Phase correlation for motion correction, to include translation, rotation, and scale-invariance. | Reddy and Chatterji 1996 |
| 2 | Turboreg | 1998 | Motion correction. | • Motion correction. • http://bigwww.epfl.ch/thevenaz/turboreg/ | Thevenaz et al. 1998 |
| 3 | subPixelPhase | 2002 | Motion correction. | • Closed-form solution to subpixel translation estimation using phase correlation. | Foroosh et al. 2002 |
| 4 | ROI | 2005 | Cell extraction | • Matrix multiplication; in some methods neuropil/background subtraction implemented. | Kerr et al. 2005; Kuchibhotla et al. 2014; Peron et al. 2015 |
| 5 | CellProfiler | 2006 | Cell segmentation | • Multi-algorithm pipeline for cell segmentation. • https://cellprofiler.org | Carpenter et al. 2006; McQuin et al. 2018; Lamprecht et al. 2007 |
| 6 | PCA-ICA | 2009 | Cell extraction | • Cell extraction using principal component analysis (PCA) followed by independent component analysis (ICA). | Mukamel et al. 2009 |
| 7 | ANTs | 2009 | Image analysis | • Suite of tools for registering and analyzing imaging data. • http://stnava.github.io/ANTs/ | Avants et al. 2009 |
| 8 | elastix | 2009 | Motion correction | • A general toolbox for rigid and non-rigid image registration. • https://elastix.lumc.nl | Klein et al. 2009 |
| 9 | Lucas–Kanade framework | 2009 | Motion correction | • Lucas-Kanade framework for non-uniform motion image registration. | Greenberg and Kerr 2009 |
| 10 | CIRF (calcium-behavior) | 2011 | Cell extraction | • Regressive model to obtain Ca2+ signal based on behavior. | Miri et al. 2011 |
| 11 | openBIS | 2011 | Data handling | • FAIR data management. • https://openbis.ch | Bauch et al. 2011 |
| 12 | Automated ROI analysis | 2012 | Cell extraction | • Automatic ellipses based ROI detection. | Francis et al. 2012 |
| 13 | OMERO | 2012 | Data handling | • Microscopy data handling. • https://www.openmicroscopy.org | Allan et al. 2012 |
| 14 | ADINA | 2013 | Cell extraction | • Sparse dictionary learning. | Diego et al. 2013 |
| 15 | TPP | 2013 | Analysis pipeline | • Tool for processing two-photon calcium imaging data, e.g. finding cells with SeNeCA. • http://uemweb.biomed.cas.cz/tpp/ | Tomek et al. 2013 |
| 16 | NMF | 2014 | Cell extraction | • Cell extraction using nonnegative matrix factorization (NMF). Followed by CNMF. | Pnevmatikakis et al. 2014; Maruyama et al. 2014 |
| 17 | SIMA | 2014 | Analysis pipeline | • Normalized cut segmentation, motion correction, etc. • https://github.com/losonczylab/sima | Kaifosh et al. 2014 |
| 18 | DataJoint | 2015 | Data handling | • Schema for data handling. • https://github.com/datajoint/datajoint-matlab | Yatsenko et al. 2015 |
| 19 | NWB | 2015 | Data handling | • Neurodata Without Borders (NWB) initiative to produce a common data format for electrophysiology and imaging studies. • https://github.com/NeurodataWithoutBorders | Teeters et al. 2015 |
| 20 | Suite2p | 2016 | Cell extraction | • Generative model along with GUIs. | Pachitariu et al. 2016 |
| 21 | CNMF (CaImAn) | 2016 | Cell extraction | • Constrained NMF (CNMF). • https://github.com/flatironinstitute/CaImAn-MATLAB | Pnevmatikakis et al. 2016 |
| 22 | CNMF-E | 2016 | Cell extraction | • CNMF + background model to handle one-photon data. • https://github.com/zhoupc/CNMF_E | Zhou et al. 2016, 2018 |
| 23 | Apthorpe CNN | 2016 | Cell segmentation | • Convolutional neural network (CNN). | Apthorpe et al. 2016 |
| 24 | moco | 2016 | Motion correction | • Fourier-transform based motion correction. • https://github.com/NTCColumbia/moco | Dubbs et al. 2016 |
| 25 | Cytomine | 2016 | Analysis GUI | • Analysis of large-scale imaging data. • https://cytomine.be | Marée et al. 2016 |
| 26 | ROI clustering | 2016 | Cell extraction | • Select high-intensity pixels then perform clustering to segment. • https://www.bu.edu/hanlab/files/2016/02/pfgc.zip | Mohammed et al. 2016 |
| 27 | CELLMax (conference) | 2017 | Cell extraction | • Cell segmentation and activity trace extraction using a maximum likelihood approach. | Ahanonu et al. 2018, 2017; Ahanonu 2018 |
| 28 | sc-CNMF | 2017 | Cell extraction | • CNMF + GMM/RNN seed cleansing. | Lu et al. 2017 |
| 29 | OASIS | 2017 | Trace analysis | • Generalized pool adjacent violators algorithm. • https://github.com/zhoupc/OASIS_matlab | Friedrich et al. 2017 |
| 30 | ABLE | 2017 | Cell segmentation | • Multiple active contours and a cost function to identify cells in 2P data. • https://github.com/StephanieRey/ABLE | Reynolds et al. 2017 |
| 31 | SCALPEL | 2017 | Cell extraction | • Dictionary learning, dissimilarity, and clustering. • https://cran.r-project.org/web/packages/scalpel/index.html | Petersen et al. 2017 |
| 32 | HNCcorr | 2017 | Cell segmentation | • Combinatorial optimization (correlation space analysis). • https://github.com/hochbaumGroup/HNCcorr | Spaen et al. 2017 |
| 33 | OnACID | 2017 | Cell extraction (online) | • NMF variant for online Ca2+ imaging processing. | Giovannucci et al. 2017 |
| 34 | EXTRACT | 2017 | Cell extraction | • Robust statistical estimation. | Inan et al. 2017 |
| 35 | NETCAL | 2017 | Analysis pipeline | • Calcium imaging analysis GUI. • https://github.com/orlandi/netcal | Orlandi et al. |
| 36 | NoRMCorre | 2017 | Motion correction. | • Piecewise rigid motion correction. • https://github.com/simonsfoundation/NoRMCorre | Pnevmatikakis and Giovannucci 2017 |
| 37 | CellReg | 2017 | Cross-session alignment | • Alignment of cells across days using a probabilistic approach. • https://github.com/zivlab/CellReg | Sheintuch et al. 2017 |
| 38 | NeuroSeg | 2017 | Cell segmentation | • Filtering and seed/clustering based cell segmentation. • https://github.com/baidatong/NeuroSeg | Guan et al. 2018 |
| 39 | CNMF-E+ | 2017 | Cell extraction | • Shrinkage estimation to improve CNMF-E initialization. | Takekawa et al. 2017 |
| 40 | Toolbox-Romano | 2017 | Analysis pipeline | • Full analysis pipeline with ROI-based segmentation • https://github.com/zebrain-lab/Toolbox-Romano-et-al | Romano et al. 2017 |
| 41 | SamuROI | 2017 | Analysis GUI | • GUI for data visualization • https://github.com/samuroi/SamuROI | Rueckl et al. 2017 |
| 42 | KNIME | 2017 | Analysis pipeline | • Workflow manager for data analysis. • https://www.knime.com | Fillbrunn et al. 2017 |
| 43 | U-Net2DS | 2017 | Cell segmentation | • Evaluated several deep learning models on Neurofinder, U-Net2DS best. • https://github.com/alexklibisz/deep-calcium | Klibisz et al. 2017 |
| 44 | CLEAN (conference) | 2018 | Cell sorting | • Machine learning based cell sorting of cell extraction outputs based on image and activity trace features. | Ahanonu et al. 2018; Ahanonu 2018 |
| 45 | FISSA | 2018 | Trace analysis | • Neuropil decontamination using local region around cell. • https://github.com/rochefort-lab/fissa | Keemink et al. 2018 |
| 46 | LSSC | 2018 | Cell segmentation | • Spectral clustering; variant to find local subset of eigenvectors. | Mishne et al. 2018 |
| 47 | PMD - PCA | 2018 | Denoising | • Spatially-localized penalized matrix decomposition for denoising; compression; and improved demixing. • https://github.com/paninski-lab/funimag | Buchanan et al. 2018 |
| 48 | MIN1PIPE | 2018 | Analysis pipeline | • Pre-processing to enhance neural signals then sc-CNMF for cell extraction. | Lu et al. 2018 |
| 49 | CaImAn (preprint) | 2018 | Analysis pipeline | • CNMF + several other processing tools. | Giovannucci et al. 2018 |
| 50 | SEUDO (preprint) | 2018 | Trace analysis | • Mixture of Gaussians + maximum likelihood; post-hoc activity trace correction. | Gauthier et al. 2018 |
| 51 | ACSAT | 2018 | Cell segmentation | • Global and local adaptive thresholding to identify neurons. • https://github.com/sshen8/acsat | Shen et al. 2018 |
| 52 | onlineMotionCorrection | 2018 | Motion correction | • Tested multiple algorithms and developed an online motion correction pipeline. • https://github.com/amitani/onlineMotionCorrection | Mitani and Komiyama 2018 |
| 53 | CIAtah | 2019 | Analysis pipeline | • 1P and 2P Imaging analysis pipeline supporting PCA-ICA, CNMF, CELLMax, EXTRACT, etc. • https://github.com/bahanonu/ciatah | Corder et al. 2019; Ahanonu 2018; Ahanonu and Corder 2022 |
| 54 | NAOMi (bioRxiv) | 2019 | Simulator | • Generative model for creating simulated calcium imaging movies. | Charles et al. 2019 |
| 55 | CALIMA | 2019 | Analysis pipeline | • Calcium imaging analysis GUI. | Radstake et al. 2019 |
| 56 | STNeuroNet | 2019 | Cell segmentation | • Convolutional neural network to detect and segment cells. | Soltanian-Zadeh et al. 2019 |
| 57 | AQuA | 2019 | Cell extraction | • Astrocyte imaging focused. Non-ROI cluster and propagation based detection of events. | Wang et al. 2019 |
| 58 | CaImAn | 2019 | Analysis pipeline | • Popular calcium imaging pipeline that includes CNMF + several other processing tools. • https://github.com/flatironinstitute/CaImAn | Giovannucci et al. 2019 |
| 59 | DL+RWL1-SF | 2019 | Cell extraction | • Dictionary learning and spatial correlation based cell extraction. | Mishne and Charles 2019 |
| 60 | Segment2P | 2019 | Cell segmentation | • Pre-process images and run through DeepLabV3. • https://github.com/NoahDolev/Segment2P | Dolev et al. 2019 |
| 61 | LANMC | 2019 | Motion correction | • Long short-term memory non-rigid motion correction, reduce computational cost by predicting non-rigid motion. | Chen et al. 2019 |
| 62 | marked point processes | 2020 | Cell extraction | • Probabilistic generative model, specifically a marked point process, to extract activity traces. | Shibue and Komaki 2020 |
| 63 | LocaNMF | 2020 | Region extraction | • Localized semi-nonnegative matrix factorization for extracting active regions. • https://github.com/ikinsella/locaNMF | Saxena et al. 2020 |
| 64 | EZcalcium | 2020 | Analysis pipeline | • Calcium imaging analysis toolbox. • https://github.com/porteralab/EZcalcium | Cantu et al. 2020 |
| 65 | OnACID-E + ring CNN | 2020 | Cell extraction (online) | • OnACID for miniscope and new ring CNN background model to improve accuracy. • https://github.com/flatironinstitute/CaImAn | Friedrich et al. 2020 |
| 66 | Auto CNMF-E sorting | 2020 | Cell sorting | • Machine learning (AutoML) based curation of CNMF-E outputs. • https://github.com/jf-lab/cnmfe-reviewer | Tran et al. 2020a,b |
| 67 | DeepInterpolation | 2020 | Denoising | • Encoder-decoder architecture with 2D conv. to denoise imaging data. • https://github.com/AllenInstitute/deepinterpolation | Lecoq et al. 2020 |
| 68 | BIAFLOWS | 2020 | Benchmarking | • Framework for benchmarking imaging analysis workflows. • https://biaflows.neubias.org | Rubens et al. 2020 |
| 69 | FIBSI | 2020 | Trace analysis | • Extension of Ramer-Douglas-Peucker algorithm to identify baseline that is used for signal detection. • https://github.com/rmcassidy/FIBSI_program | Cassidy et al. 2020; Alles et al. 2021 |
| 70 | DISCo | 2020 | Cell segmentation | • Pixel correlation and deep learning (CNN) + graph based segmentation. • https://github.com/EKirschbaum/DISCo | Kirschbaum et al. 2020 |
| 71 | DeepCINAC | 2020 | Trace analysis | • Trace analysis after human labeling followed by CNNs + bidirectional long-short term memory (LSTM) network. • https://gitlab.com/cossartlab/deepcinac | Denis et al. 2020 |
| 72 | NDSEP | 2020 | Cell extraction | • Dataflow framework for real-time calcium imaging processing. • http://dspcad-www.iacs.umd.edu/bcnm/index.html | Lee et al. 2020 |
| 73 | DeepBrainSeg | 2020 | Segmentation | • Dual-pathway CNN to learn local and contextual features. | Tan et al. 2020 |
| 74 | RT-3DMC | 2020 | Motion correction | • Bead or soma tracking for real-time motion correction during 2P imaging. • https://github.com/SilverLabUCL/SilverLab-Microscope | Griffiths et al. 2020 |
| 75 | Cellpose | 2021 | Cell segmentation | • Neural network and gradient-based cell segmentation. • https://github.com/mouseland/cellpose | Stringer et al. 2021 |
| 76 | NAOMi | 2021 | Simulator | • Detailed model simulation for benchmarking calcium imaging algorithms. • https://bitbucket.org/adamshch/naomi_sim/src/master/ | Song et al. 2021 |
| 77 | OnACID-E + ring CNN | 2021 | Cell extraction (online) | • OnACID for 1P data and ring CNN background model. • https://github.com/flatironinstitute/CaImAn | Friedrich et al. 2021 |
| 78 | EXTRACT | 2021 | Cell extraction | • Robust statistics based cell extraction. • https://github.com/schnitzer-lab/EXTRACT-public | Inan et al. 2021 |
| 79 | Minian | 2021 | Analysis pipeline | • Imaging analysis pipeline with CNMF for cell extraction, in part using Jupyter notebooks with GUI elements. • https://github.com/DeniseCaiLab/minian | Dong et al. 2021 |
| 80 | Mesmerize | 2021 | Analysis pipeline | • Imaging analysis platform with CaImAn for cell extraction, import support for other cell extraction algorithms. • https://github.com/kushalkolar/MESmerize | Kolar et al. 2021 |
| 81 | DeepInterpolation | 2021 | Denoising | • Encoder-decoder architecture with 2D conv. to denoise imaging data. • https://github.com/AllenInstitute/deepinterpolation | Lecoq et al. 2021 |
| 82 | BEAR | 2021 | Cell extraction | • Neural network approximation of PCA for cell extraction. • https://github.com/NICALab/BEAR | Han et al. 2021 |
| 83 | CaPTure | 2021 | Cell extraction | • ROI segmentation and activity extraction. • https://github.com/LieberInstitute/CaPTure | Tippani et al. 2021 |
| 84 | CASCADE | 2021 | Trace analysis | • Spike inference based on dual ephys/calcium imaging recordings. • https://github.com/HelmchenLabSoftware/Cascade | Rupprecht et al. 2021 |
| 85 | VolPy | 2021 | Analysis pipeline | • Voltage imaging analysis pipeline integrated into CaImAn. • https://github.com/flatironinstitute/CaImAn | Cai et al. 2021 |
| 86 | DeepCAD | 2021 | Denoising | • Deep neural network based denoising. • https://github.com/cabooster/DeepCAD-RT | Li et al. 2021 |
| 87 | SpecSeg | 2021 | Cell extraction | • Spectral density of pixels to identify ROIs. Also incorporates motion correction and cross-session matching. • https://github.com/Leveltlab/SpectralSegmentation | de Kraker et al. 2021 |
| 88 | FIOLA | 2021 | Cell extraction (online) | • GPU- and computational graph-based speed-ups along with non-negative least squares for post-initialization signal extraction. • https://github.com/nel-lab/FIOLA | Giovannucci et al. 2021 |
| 89 | PatchWarp | 2021 | Motion correction | • Affine transformation of subfields followed by stitching subfields together. • https://github.com/ryhattori/PatchWarp | Hattori and Komiyama 2021 |
| 90 | MVG-CNN | 2021 | Region extraction | • Automated sleep states classification using multiplex visibility graphs and deep learning. Data URL. • https://github.com/comp-imaging-sci/MVG-CNN | Zhang et al. 2021 |
| 91 | Flow-Registration | 2021 | Motion correction | • Variational optical flow for non-uniform motion correction • https://github.com/phflot/flow_registration | Flotho et al. 2022 |
| 92 | SUNS | 2021 | Cell segmentation | • Cell segmentation using shallow U-Nets. • https://github.com/YijunBao/Shallow-UNet-Neuron-Segmentation_SUNS | Bao et al. 2021 |
| 93 | Carignan | 2021 | Cell extraction | • Online cell extraction and triggering based on OnACID and CaImAn. • https://github.com/tzklab/carignan | Taniguchi et al. 2021 |
| 94 | MullenClassifier | 2021 | Cell sorting | • Feature extraction from cell images and tracs followed by supervised learning classifier. | Mullen et al. 2021 |
| 95 | timeUnet | 2021 | Denoising | • Deep learning for denoising with temporal information added in. • https://github.com/BoHuangLab/Transfer-Learning-Denoising/ | Wang et al. 2021 |
| 96 | EMC2 | 2021 | Motion correction | • Wavelet decomposition to detect bright spots followed by motion correction with multiple hypothesis tracking and computing elastic deformation. • https://icy.bioimageanalysis.org/plugin/elastic-motion-correction-concatenation-emc2-of-tracks/ | Lagache et al. 2021 |
| 97 | GraFT | 2022 | Cell extraction | • Dictionary-based learning of activity traces followed by graph-based segmentation. • https://github.com/adamshch/GraFT-analysis | Charles et al. 2022 |
| 98 | CaPTure | 2022 | Analysis pipeline | • Binary/watershed segmentation followed by ROI-based mean traces. • https://github.com/LieberInstitute/CaPTure | Tippani et al. 2022 |
| 99 | SpecSeg | 2022 | Cell segmentation | • Cross spectral power-based segmentation of neurons and neurites. • https://github.com/Leveltlab/SpectralSegmentation | de Kraker et al. 2022 |
| 100 | CITE-On | 2022 | Cell extraction | • Online cell detection and trace extraction using CNNs. • https://gitlab.iit.it/fellin-public/cite-on | Sità et al. 2022 |
| 101 | DL-assisted 2P fiberscope | 2022 | Denoising | • Denoising 2P fiberscope data using deep neural network (conditional generative adversarial network). • https://figshare.com/articles/dataset/Data/19193792 | Guan et al. 2022 |
| 102 | 4SM | 2022 | Cell extraction | • Generative adversarial network for image segmentation. • https://github.com/SharifAmit/4SM | Kamran et al. 2022 |
| 103 | DeepCAD-RT | 2022 | Denoising | • Improved version of DeepCAD for real time performance. • https://github.com/cabooster/DeepCAD-RT/ | Li et al. 2023a |
| 104 | SEUDO | 2022 | Trace analysis | • Mixture of Gaussians + maximum likelihood; post-hoc activity trace correction. • https://github.com/adamshch/SEUDO | Gauthier et al. 2022 |
| 105 | AxialMotionCorrect | 2022 | Motion correction | • Axial motion correction via multi-plane scanning plus maximum likelihood optimization. • https://gitlab.com/anflores/axial_motion_correction | Flores-Valle and Seelig 2022 |
| 106 | FIFER | 2022 | Motion correction | • Feature-based motion correction, finding features using a density-based estimating and clustering algorithm and matching features with a similarity metric for registration. • https://github.com/Weiyi-Liu-Unique/FIFER | Liu et al. 2022 |
| 107 | NWB | 2022 | Data handling | • Neurodata Without Borders (NWB) to standardize ephys and imaging data across tools. • https://github.com/NeurodataWithoutBorders | Rübel et al. 2022 |
| 108 | DeCalciOn | 2023 | Online analysis pipeline | • Integrate hardware and software to online decode calcium signals. • https://github.com/zhe-ch/ACTEV | Chen et al. 2023 |
| 109 | jGCaMP8 | 2023 | Calcium indicator | • Improved calcium indicators with increased sensitivity and reduced background. | Zhang et al. 2023a |
| 110 | NeuroSeg-II | 2023 | Cell segmentation | • 2P cell segmentation using region-based convolutional neural network with modifications. • https://github.com/XZH-James/NeuroSeg2 | Xu et al. 2023 |
| 111 | CaliAli | 2023 | Cross-session alignment | • Cross-session alignment using vasculature information. • https://github.com/CaliAli-PV/CaliAli | Vergara et al. 2023 |
| 112 | DeepWonder | 2023 | Cell extraction | • Deep-learning-based cell finding for widefield datasets. • https://github.com/yuanlong-o/Deep_widefield_cal_inferece | Zhang et al. 2023b |
| 113 | ASTRA | 2023 | Cell segmentation | • Deep neural network for astrocyte segmentation. • https://gitlab.iit.it/fellin-public/astra | Bonato et al. 2023 |
| 114 | SRDTrans | 2023 | Denoise | • Spatial redundancy for training followed by spatiotemporal transformer architecture to reduce CNN bias/issues. • https://github.com/cabooster/SRDTrans | Li et al. 2023b |
| 115 | REALS | 2023 | Motion correction | • Motion correction via simultaneous transformation and low rank and sparse decomposition with gradient-based updates. • https://openaccess.thecvf.com/content/WACV2023/supplemental/Cho_Robust_and_Efficient_WACV_2023_supplemental.zip | Cho et al. 2023 |
| 116 | LD-MCM | 2023 | Motion correction | • Motion correction using deep learning feature identification and control point registration. • https://github.com/bahanonu/ciatah | Ahanonu et al. 2023 |
B Srinivasa Reddy and Biswanath N Chatterji. An fft-based technique for translation, rotation, and scale-invariant image registration. IEEE transactions on image processing, 5(8):1266–1271, 1996.
Philippe Thevenaz, Urs E Ruttimann, and Michael Unser. A pyramid approach to subpixel registration based on intensity. Image Processing, IEEE Transactions on, 7(1):27–41, 1998.
Hassan Foroosh, Josiane B Zerubia, and Marc Berthod. Extension of phase correlation to subpixel registration. IEEE transactions on image processing, 11(3):188–200, 2002.
J N Kerr, D Greenberg, and F Helmchen. Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci U S A, 102(39): 14063–14068, 2005. ISSN 0027-8424 (Print) 0027-8424. doi: 10.1073/pnas. 0506029102.
K V Kuchibhotla, S Wegmann, K J Kopeikina, J Hawkes, N Rudinskiy, M L Andermann, T L Spires-Jones, B J Bacskai, and B T Hyman. Neurofibrillary tangle-bearing neurons are functionally integrated in cortical circuits in vivo. Proc Natl Acad Sci U S A, 111(1):510–514, 2014. ISSN 0027-8424. doi: 10.1073/pnas.1318807111.
Simon P. Peron, Jeremy Freeman, Vijay Iyer, Caiying Guo, and Karel Svoboda. A Cellular Resolution Map of Barrel Cortex Activity during Tactile Behavior. Neuron, 86(3): 783–799, 2015. ISSN 10974199. doi: 10.1016/j.neuron.2015.03.027. URL http://dx.doi.org/10.1016/j.neuron.2015.03.027.
Anne E Carpenter, Thouis R Jones, Michael R Lamprecht, Colin Clarke, In Han Kang, Ola Friman, David A Guertin, Joo Han Chang, Robert A Lindquist, Jason Moffat, et al. Cellprofiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology, 7(10):1–11, 2006.
Claire McQuin, Allen Goodman, Vasiliy Chernyshev, Lee Kamentsky, Beth A Cimini, Kyle W Karhohs, Minh Doan, Liya Ding, Susanne M Rafelski, Derek Thirstrup, et al. Cellprofiler 3.0: Next-generation image processing for biology. PLoS biology, 16(7):e2005970, 2018.
Michael R Lamprecht, David M Sabatini, and Anne E Carpenter. Cellprofiler™: free, versatile software for automated biological image analysis. Biotechniques, 42(1):71–75, 2007.
Eran A Mukamel, Axel Nimmerjahn, and Mark J Schnitzer. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron, 63(6):747–760, 2009.
Brian B Avants, Nick Tustison, Gang Song, et al. Advanced normalization tools (ants). Insight j, 2(365):1–35, 2009.
Stefan Klein, Marius Staring, Keelin Murphy, Max A Viergever, and
Josien PW Pluim. Elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging, 29(1):196–205, 2009.
David S Greenberg and Jason ND Kerr. Automated correction of fast motion artifacts for two-photon imaging of awake animals. Journal of neuroscience methods, 176(1):1–15, 2009.
A Miri, K Daie, R D Burdine, E Aksay, and D W Tank. Regression-based identification of behavior-encoding neurons during large-scale optical imaging of neural activity at cellular resolution. J Neurophysiol, 105(2):964–980, 2011. ISSN 1522-1598 (Electronic) 0022-3077 (Linking). doi: 10.1152/jn.00702.2010. URL http://www.ncbi.nlm.nih.gov/pubmed/21084686.
Angela Bauch, Izabela Adamczyk, Piotr Buczek, Franz-Josef Elmer, Kaloyan Enimanev, Pawel Glyzewski, Manuel Kohler, Tomasz Pylak, Andreas Quandt, Chandrasekhar Ramakrishnan, et al. openbis: a flexible framework for managing and analyzing complex data in biology research. BMC bioinformatics, 12(1):1–19, 2011.
M Francis, X Qian, C Charbel, J Ledoux, J C Parker, and M S Taylor. Automated region of interest analysis of dynamic Ca(2)+ signals in image sequences. Am J Physiol Cell Physiol, 303(3):C236–43, 2012. ISSN 0363-6143. doi: 10.1152/ajpcell.00016.2012.
Chris Allan, Jean-Marie Burel, Josh Moore, Colin Blackburn, Melissa Linkert, Scott Loynton, Donald MacDonald, William J Moore, Carlos Neves, Andrew Patterson, et al. Omero: flexible, model-driven data management for experimental biology. Nature methods, 9(3):245–253, 2012.
Ferran Diego, Susanne Reichinnek, Martin Both, and Fred A Hamprecht.
Automated identification of neuronal activity from calcium imaging by sparse dictionary learning. In Biomedical Imaging (ISBI), 2013 IEEE 10th International Symposium on, pages 1058–1061. IEEE, 2013.
Jakub Tomek, Ondrej Novak, and Josef Syka. Two-photon processor and seneca: a freely available software package to process data from two-photon calcium imaging at speeds down to several milliseconds per frame. Journal of neurophysiology, 110(1):243–256, 2013.
Eftychios A Pnevmatikakis, Yuanjun Gao, Daniel Soudry, David Pfau, Clay Lacefield, Kira Poskanzer, Randy Bruno, Rafael Yuste, and Liam Paninski. A structured matrix factorization framework for large scale calcium imaging data analysis. arXiv preprint arXiv:1409.2903, 2014.
Ryuichi Maruyama, Kazuma Maeda, Hajime Moroda, Ichiro Kato, Masashi Inoue, Hiroyoshi Miyakawa, and Toru Aonishi. Detecting cells using non-negative matrix factorization on calcium imaging data. Neural Netw, 55:11–19, mar 2014. ISSN 0893-6080. doi: 10.1016/j.neunet.2014. 03.007. URL http://www.ncbi.nlm.nih.gov/pubmed/24705544.
Patrick Kaifosh, Jeffrey D Zaremba, Nathan B Danielson, and Attila Losonczy. SIMA: Python software for analysis of dynamic fluorescence imaging data. Frontiers in neuroinformatics, 8:80, 2014.
Dimitri Yatsenko, Jacob Reimer, Alexander S Ecker, Edgar Y Walker, Fabian Sinz, Philipp Berens, Andreas Hoenselaar, R James Cotton, Athanassios S Siapas, and Andreas S Tolias. Datajoint: managing big scientific data using matlab or python. BioRxiv, page 031658, 2015.
Jeffery L Teeters, Keith Godfrey, Rob Young, Chinh Dang, Claudia Friedsam, Barry Wark, Hiroki Asari, Simon Peron, Nuo Li, and Adrien
Peyrache. Neurodata without borders: creating a common data format for neurophysiology. Neuron, 88(4):629–634, 2015.
Marius Pachitariu, Carsen Stringer, Sylvia Schröder, Mario Dipoppa, L Federico Rossi, Matteo Carandini, and Kenneth D Harris. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. Biorxiv, page 061507, 2016.
Eftychios A Pnevmatikakis, Daniel Soudry, Yuanjun Gao, Timothy A Machado, Josh Merel, David Pfau, Thomas Reardon, Yu Mu, Clay Lacefield, Weijian Yang, et al. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron, 89(2):285–299, 2016.
P Zhou, SL Resendez, GD Stuber, RE Kass, and L Paninski. Efficient and accurate extraction of in vivo calcium signals from microendoscope video data. arXiv preprint arXiv:1605.07266, 2016.
Pengcheng Zhou, Shanna L Resendez, Jose Rodriguez-Romaguera, Jessica C Jimenez, Shay Q Neufeld, Andrea Giovannucci, Johannes Friedrich, Eftychios A Pnevmatikakis, Garret D Stuber, Rene Hen, et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. ELife, 7:e28728, 2018.
Noah Apthorpe, Alexander Riordan, Robert Aguilar, Jan Homann, Yi Gu, David Tank, and H Sebastian Seung. Automatic neuron detection in calcium imaging data using convolutional networks. In Advances in Neural Information Processing Systems, pages 3270–3278, 2016.
Alexander Dubbs, James Guevara, and Rafael Yuste. moco: Fast motion correction for calcium imaging. Frontiers in neuroinformatics, 10:6, 2016.
Raphaël Marée, Loïc Rollus, Benjamin Stévens, Renaud Hoyoux, Gilles Louppe, Rémy Vandaele, Jean-Michel Begon, Philipp Kainz, Pierre Geurts, and Louis Wehenkel. Collaborative analysis of multi-gigapixel imaging data using cytomine. Bioinformatics, 32(9):1395–1401, 2016.
Ali I Mohammed, Howard J Gritton, Hua-an Tseng, Mark E Bucklin, Zhaojie Yao, and Xue Han. An integrative approach for analyzing hundreds of neurons in task performing mice using wide-field calcium imaging. Scientific reports, 6(1):20986, 2016.
B. Ahanonu, L. J. Kitch, T. H. Kim, M. C. Larkin, E. O. Hamel, J. Lecoq, D. E. Aldarondo, and M. J. Schnitzer. Maximum likelihood and machine learning based methods for automated cell sorting of large-scale neural calcium imaging data. Society for Neuroscience, 2018. URL https://abstractsonline.com/pp8/#!/4649/presentation/41917.
B. Ahanonu, L. J. Kitch, T. H. Kim, M. C. Larkin, E. O. Hamel, J. Lecoq, and M. J. Schnitzer. Maximum likelihood based cell sorting of large-scale neural calcium imaging data. Society for Neuroscience, 2017. URL http://www.abstractsonline.com/pp8/index.html#!/4376/presentation/18520.
Biafra Owowonta Ahanonu. Neural Ensemble Dynamics in Behaving Animals: Computational Approaches and Applications in Amygdala and Striatum. Stanford University, 2018.
Jinghao Lu, Chunyuan Li, and Fan Wang. Seeds cleansing cnmf for spatiotemporal neural signals extraction of miniscope imaging data. arXiv preprint arXiv:1704.00793, 2017.
Johannes Friedrich, Pengcheng Zhou, and Liam Paninski. Fast online deconvolution of calcium imaging data. PLoS computational biology, 13 (3):e1005423, 2017.
Stephanie Reynolds, Therese Abrahamsson, Renaud Schuck, P Jesper Sjöström, Simon R Schultz, and Pier Luigi Dragotti. Able: An activity-based level set segmentation algorithm for two-photon calcium imaging data. eNeuro, pages ENEURO–0012, 2017.
Ashley Petersen, Noah Simon, and Daniela Witten. SCALPEL: Extracting Neurons from Calcium Imaging Data. ArXiv e-prints, art. arXiv:1703.06946, March 2017.
Quico Spaen, Dorit S Hochbaum, and Roberto Asín-Achá. Hnccorr: A novel combinatorial approach for cell identification in calcium-imaging movies. arXiv preprint arXiv:1703.01999, 2017.
Andrea Giovannucci, Johannes Friedrich, Matt Kaufman, Anne Churchland, Dmitri Chklovskii, Liam Paninski, and Eftychios A Pnevmatikakis. Onacid: Online analysis of calcium imaging data in real time. In Advances in Neural Information Processing Systems, pages 2381–2391, 2017.
Hakan Inan, Murat A Erdogdu, and Mark Schnitzer. Robust estimation of neural signals in calcium imaging. In Advances in Neural Information Processing Systems, pages 2901–2910, 2017.
JG Orlandi, S Fernández-García, A Comella-Bolla, M Masana, G García-Díaz Barriga, M Yaghoubi, A Kipp, JM Canals, MA Colicos, J Davidsen,
et al. Netcal: An interactive platform for large-scale, network and population dynamics analysis of calcium imaging recordings, zenodo (2017).
Eftychios A Pnevmatikakis and Andrea Giovannucci. Normcorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. Journal of neuroscience methods, 291:83–94, 2017.
Liron Sheintuch, Alon Rubin, Noa Brande-Eilat, Nitzan Geva, Noa Sadeh, Or Pinchasof, and Yaniv Ziv. Tracking the same neurons across multiple days in ca2+ imaging data. Cell reports, 21(4):1102–1115, 2017.
Jiangheng Guan, Jingcheng Li, Shanshan Liang, Ruijie Li, Xingyi Li, Xiaozhe Shi, Ciyu Huang, Jianxiong Zhang, Junxia Pan, Hongbo Jia, et al. Neuroseg: automated cell detection and segmentation for in vivo two-photon ca 2+ imaging data. Brain Structure and Function, 223(1): 519–533, 2018.
Takashi Takekawa, Hirotaka Asai, Noriaki Ohkawa, Masanori Nomoto, Reiko Okubo-Suzuki, Khaled Ghandour, Masaaki Sato, Yasunori Hayashi, Kaoru Inokuchi, and Tomoki Fukai. Automatic sorting system for large calcium imaging data. bioRxiv, page 215145, 2017.
Sebastián A Romano, Verónica Pérez-Schuster, Adrien Jouary, Jonathan Boulanger-Weill, Alessia Candeo, Thomas Pietri, and Germán Sumbre. An integrated calcium imaging processing toolbox for the analysis of neuronal population dynamics. PLoS computational biology, 13(6): e1005526, 2017.
Martin Rueckl, Stephen C Lenzi, Laura Moreno-Velasquez, Daniel Parthier, Dietmar Schmitz, Sten Ruediger, and Friedrich W Johenning.
Samuroi, a python-based software tool for visualization and analysis of dynamic time series imaging at multiple spatial scales. Frontiers in neuroinformatics, 11:44, 2017.
Alexander Fillbrunn, Christian Dietz, Julianus Pfeuffer, René Rahn, Gregory A Landrum, and Michael R Berthold. Knime for reproducible cross-domain analysis of life science data. Journal of biotechnology, 261: 149–156, 2017.
Aleksander Klibisz, Derek Rose, Matthew Eicholtz, Jay Blundon, and Stanislav Zakharenko. Fast, simple calcium imaging segmentation with fully convolutional networks. In International Workshop on Deep Learning in Medical Image Analysis, pages 285–293. Springer, 2017.
Sander W Keemink, Scott C Lowe, Janelle MP Pakan, Evelyn Dylda, Mark CW Van Rossum, and Nathalie L Rochefort. Fissa: A neuropil decontamination toolbox for calcium imaging signals. Scientific reports, 8 (1):1–12, 2018.
Gal Mishne, Ronald R Coifman, Maria Lavzin, and Jackie Schiller. Automated cellular structure extraction in biological images with applications to calcium imaging data. bioRxiv, page 313981, 2018.
E Kelly Buchanan, Ian Kinsella, Ding Zhou, Rong Zhu, Pengcheng Zhou, Felipe Gerhard, John Ferrante, Ying Ma, Sharon Kim, Mohammed Shaik, et al. Penalized matrix decomposition for denoising, compression, and improved demixing of functional imaging data. bioRxiv, page 334706, 2018.
Jinghao Lu, Chunyuan Li, Jonnathan Singh-Alvarado, Zhe Charles Zhou, Flavio Fröhlich, Richard Mooney, and Fan Wang. MIN1PIPE: A
Miniscope 1-Photon-Based Calcium Imaging Signal Extraction Pipeline. Cell Reports, 23(12):3673–3684, 2018. ISSN 2211-1247.
Andrea Giovannucci, Johannes Friedrich, Pat Gunn, Jeremie Kalfon, Sue Ann Koay, Jiannis Taxidis, Farzaneh Najafi, Jeffrey L Gauthier, Pengcheng Zhou, and David W Tank. CaImAn: An open source tool for scalable Calcium Imaging data Analysis. bioRxiv, page 339564, 2018.
Jeffrey L Gauthier, Sue Ann Koay, Edward H Nieh, David W Tank, Jonathan W Pillow, and Adam S Charles. Detecting and correcting false transients in calcium imaging. bioRxiv, page 473470, 2018.
Simon P Shen, Hua-an Tseng, Kyle R Hansen, Ruofan Wu, Howard J Gritton, Jennie Si, and Xue Han. Automatic cell segmentation by adaptive thresholding (acsat) for large-scale calcium imaging datasets. eneuro, 5(5), 2018.
Akinori Mitani and Takaki Komiyama. Real-time processing of two-photon calcium imaging data including lateral motion artifact correction. Frontiers in neuroinformatics, 12:98, 2018.
Gregory Corder, Biafra Ahanonu, Benjamin F Grewe, Dong Wang, Mark J Schnitzer, and Grégory Scherrer. An amygdalar neural ensemble that encodes the unpleasantness of pain. Science, 363(6424):276–281, 2019.
Biafra Ahanonu and Gregory Corder. Recording pain-related brain activity in behaving animals using calcium imaging calcium imaging and miniature microscopes. In Contemporary Approaches to the Study of Pain: From Molecules to Neural Networks, pages 217–276. Springer, 2022.
Adam S Charles, Alex Song, Jeffrey L Gauthier, Jonathan W Pillow, and David W Tank. Neural anatomy and optical microscopy (naomi) simulation for evaluating calcium imaging methods. bioRxiv, page 726174, 2019.
FDW Radstake, EAL Raaijmakers, R Luttge, Svitlana Zinger, and Jean-Philippe Frimat. Calima: The semi-automated open-source calcium imaging analyzer. Computer methods and programs in biomedicine, 179: 104991, 2019.
Somayyeh Soltanian-Zadeh, Kaan Sahingur, Sarah Blau, Yiyang Gong, and Sina Farsiu. Fast and robust active neuron segmentation in two-photon calcium imaging using spatiotemporal deep learning. Proceedings of the National Academy of Sciences, 116(17):8554–8563, 2019.
Yizhi Wang, Nicole V DelRosso, Trisha V Vaidyanathan, Michelle K Cahill, Michael E Reitman, Silvia Pittolo, Xuelong Mi, Guoqiang Yu, and Kira E Poskanzer. Accurate quantification of astrocyte and neurotransmitter fluorescence dynamics for single-cell and population-level physiology. Nature Neuroscience, 22(11):1936–1944, 2019.
Andrea Giovannucci, Johannes Friedrich, Pat Gunn, Jeremie Kalfon, Brandon L Brown, Sue Ann Koay, Jiannis Taxidis, Farzaneh Najafi, Jeffrey L Gauthier, Pengcheng Zhou, et al. Caiman an open source tool for scalable calcium imaging data analysis. Elife, 8:e38173, 2019.
Gal Mishne and Adam S Charles. Learning spatially-correlated temporal dictionaries for calcium imaging. In ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pages 1065–1069. IEEE, 2019.
Noah Dolev, Lior Pinkus, and Michal Rivlin-Etzion. Segment2p: Parameter-free automated segmentation of cellular fluorescent signals. BioRxiv, page 832188, 2019.
Zhe Chen, Hugh T Blair, and Jason Cong. Lanmc: Lstm-assisted non-rigid motion correction on fpga for calcium image stabilization. In Proceedings of the 2019 ACM/SIGDA International Symposium on Field-Programmable Gate Arrays, pages 104–109, 2019.
Ryohei Shibue and Fumiyasu Komaki. Deconvolution of calcium imaging data using marked point processes. PLoS computational biology, 16(3): e1007650, 2020.
Shreya Saxena, Ian Kinsella, Simon Musall, Sharon H Kim, Jozsef Meszaros, David N Thibodeaux, Carla Kim, John Cunningham, Elizabeth MC Hillman, Anne Churchland, et al. Localized semi-nonnegative matrix factorization (locanmf) of widefield calcium imaging data. PLOS Computational Biology, 16(4):e1007791, 2020.
Daniel A Cantu, Bo Wang, Michael W Gongwer, Cynthia X He, Anubhuti Goel, Anand Suresh, Nazim Kourdougli, Erica D Arroyo, William Zeiger, and Carlos Portera-Cailliau. Ezcalcium: Open source toolbox for analysis of calcium imaging data. bioRxiv, 2020.
Johannes Friedrich, Andrea Giovannucci, and Eftychios A Pnevmatikakis. Online analysis of microendoscopic 1-photon calcium imaging data streams. bioRxiv, 2020.
Lina M Tran, Andrew J Mocle, Adam I Ramsaran, Alex D Jacob,
Paul W Frankland, and Sheena A Josselyn. Automated curation of cnmf-e-extracted roi spatial footprints and calcium traces using open-source automl tools. bioRxiv, 2020a.
Lina M Tran, Andrew J Mocle, Adam I Ramsaran, Alexander D Jacob, Paul W Frankland, and Sheena A Josselyn. Automated curation of cnmf-e-extracted roi spatial footprints and calcium traces using open-source automl tools. Frontiers in Neural Circuits, 14:42, 2020b.
Jerome Lecoq, Michael Oliver, Joshua H Siegle, Natalia Orlova, and Christof Koch. Removing independent noise in systems neuroscience data using deepinterpolation. bioRxiv, 2020.
Ulysse Rubens, Romain Mormont, Lassi Paavolainen, Volker Bäcker, Benjamin Pavie, Leandro A Scholz, Gino Michiels, Martin Maška, Devrim Ünay, Graeme Ball, et al. Biaflows: A collaborative framework to reproducibly deploy and benchmark bioimage analysis workflows. Patterns, 1(3):100040, 2020.
Ryan M Cassidy, Alexis G Bavencoffe, Elia R Lopez, Sai S Cheruvu, Edgar T Walters, Rosa A Uribe, Anne Marie Krachler, and Max A Odem. Frequency-independent biological signal identification (fibsi): A free program that simplifies intensive analysis of non-stationary time series data. bioRxiv, 2020.
Sascha RA Alles, Max A Odem, Van B Lu, Ryan M Cassidy, and Peter A Smith. Chronic bdnf simultaneously inhibits and unmasks superficial dorsal horn neuronal activity. Scientific reports, 11(1):1–14, 2021.
Elke Kirschbaum, Alberto Bailoni, and Fred A Hamprecht. Disco: deep
learning, instance segmentation, and correlations for cell segmentation in calcium imaging. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part V 23, pages 151–162. Springer, 2020.
Julien Denis, Robin F Dard, Eleonora Quiroli, Rosa Cossart, and Michel A Picardo. Deepcinac: a deep-learning-based python toolbox for inferring calcium imaging neuronal activity based on movie visualization. eneuro, 7(4), 2020.
Yaesop Lee, Jing Xie, Eungjoo Lee, Srijesh Sudarsanan, Da-Ting Lin, Rong Chen, and Shuvra S Bhattacharyya. Real-time neuron detection and neural signal extraction platform for miniature calcium imaging. Frontiers in Computational Neuroscience, 14:43, 2020.
Chaozhen Tan, Yue Guan, Zhao Feng, Hong Ni, Zoutao Zhang, Zhiguang Wang, Xiangning Li, Jing Yuan, Hui Gong, Qingming Luo, et al. Deepbrainseg: Automated brain region segmentation for micro-optical images with a convolutional neural network. Frontiers in neuroscience, 14: 179, 2020.
Victoria A Griffiths, Antoine M Valera, Joanna YN Lau, Hana Roš, Thomas J Younts, Bóris Marin, Chiara Baragli, Diccon Coyle, Geoffrey J Evans, George Konstantinou, et al. Real-time 3d movement correction for two-photon imaging in behaving animals. Nature methods, 17(7):741–748, 2020.
Carsen Stringer, Tim Wang, Michalis Michaelos, and Marius Pachitariu. Cellpose: a generalist algorithm for cellular segmentation. Nature Methods, 18(1):100–106, 2021.
Alexander Song, Jeff L Gauthier, Jonathan W Pillow, David W Tank, and Adam S Charles. Neural anatomy and optical microscopy (naomi) simulation for evaluating calcium imaging methods. Journal of Neuroscience Methods, 358:109173, 2021.
Johannes Friedrich, Andrea Giovannucci, and Eftychios A Pnevmatikakis. Online analysis of microendoscopic 1-photon calcium imaging data streams. PLoS computational biology, 17(1):e1008565, 2021.
Hakan Inan, Claudia Schmuckermair, Tugce Tasci, Biafra Ahanonu, Oscar Hernandez, Jérôme Lecoq, Fatih Dinç, Mark J Wagner, Murat Erdogdu, and Mark J Schnitzer. Fast and statistically robust cell extraction from large-scale neural calcium imaging datasets. bioRxiv, 2021.
Zhe Dong, William Mau, Yu Susie Feng, Zachary T Pennington, Lingxuan Chen, Yosif Zaki, Kanaka Rajan, Tristan Shuman, Daniel Aharoni, and Denise J Cai. Minian: An open-source miniscope analysis pipeline. bioRxiv, 2021.
Kushal Kolar, Daniel Dondorp, Jordi Cornelis Zwiggelaar, Jørgen Høyer, and Marios Chatzigeorgiou. Mesmerize: a dynamically adaptable user-friendly analysis platform for 2d & 3d calcium imaging data. bioRxiv, page 840488, 2021.
Jérôme Lecoq, Michael Oliver, Joshua H Siegle, Natalia Orlova, Peter Ledochowitsch, and Christof Koch. Removing independent noise in systems neuroscience data using deepinterpolation. Nature Methods, pages 1–8, 2021.
Seungjae Han, Eun-Seo Cho, Inkyu Park, Kijung Shin, and Young-Gyu Yoon. Efficient neural network approximation of robust pca for automated analysis of calcium imaging data. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 595–604. Springer, 2021.
Madhavi Tippani, Elizabeth A Pattie, Brittany A Davis, Claudia V Nguyen, Yanhong Wang, Srinidhi Rao Sripathy, Brady J Maher, Keri Martinowich, Andrew E Jaffe, and Stephanie Cerceo Page. Capture: Calcium peak toolbox for analysis of in vitro calcium imaging data. bioRxiv, 2021.
Peter Rupprecht, Stefano Carta, Adrian Hoffmann, Mayumi Echizen, Antonin Blot, Alex C Kwan, Yang Dan, Sonja B Hofer, Kazuo Kitamura, Fritjof Helmchen, et al. A database and deep learning toolbox for noise-optimized, generalized spike inference from calcium imaging. Nature Neuroscience, 24(9):1324–1337, 2021.
Changjia Cai, Johannes Friedrich, Amrita Singh, M Hossein Eybposh, Eftychios A Pnevmatikakis, Kaspar Podgorski, and Andrea Giovannucci. Volpy: automated and scalable analysis pipelines for voltage imaging datasets. PLoS computational biology, 17(4):e1008806, 2021.
Xinyang Li, Guoxun Zhang, Jiamin Wu, Yuanlong Zhang, Zhifeng Zhao, Xing Lin, Hui Qiao, Hao Xie, Haoqian Wang, Lu Fang, et al. Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising. Nature Methods, pages 1–6, 2021.
Leander de Kraker, Koen Seignette, Premnath Thamizharasu, Bastijn JG van den Boom, Ildefonso Ferreira Pica, Ingo Willuhn, Christiaan N Levelt, and
Chris van der Togt. Specseg: cross spectral power-based segmentation of neurons and neurites in chronic calcium imaging datasets. bioRxiv, pages 2020–10, 2021.
Andrea Giovannucci, Changjia Cai, Cynthia Dong, Marton Rozsa, and Eftychios Pnevmatikakis. Fiola: An accelerated pipeline for fluorescence imaging online analysis. 2021.
Ryoma Hattori and Takaki Komiyama. Patchwarp: Corrections of non-uniform image distortions in two-photon calcium imaging data by patchwork affine transformations. bioRxiv, 2021. doi: 10.1101/2021.11.10.468164. URL https://www.biorxiv.org/content/early/2021/11/13/2021.11.10.468164.
Xiaohui Zhang, Eric C Landsness, Wei Chen, Hanyang Miao, Michelle Tang, Lindsey M Brier, Joseph P Culver, Jin-Moo Lee, and Mark A Anastasio. Automated sleep state classification of wide-field calcium imaging data via multiplex visibility graphs and deep learning. Journal of Neuroscience Methods, page 109421, 2021.
Philipp Flotho, Shinobu Nomura, Bernd Kuhn, and Daniel J Strauss. Software for non-parametric image registration of 2-photon imaging data. Journal of Biophotonics, 15(8):e202100330, 2022.
Yijun Bao, Somayyeh Soltanian-Zadeh, Sina Farsiu, and Yiyang Gong. Segmentation of neurons from fluorescence calcium recordings beyond real time. Nature machine intelligence, 3(7):590–600, 2021.
Masaki Taniguchi, Taro Tezuka, Pablo Vergara, Sakthivel Srinivasan, Takuma Hosokawa, Yoan Chérasse, Toshie Naoi, Takeshi Sakurai, and Masanori Sakaguchi. Open-source software for real-time calcium
imaging and synchronized neuron firing detection. In 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pages 2997–3003. IEEE, 2021.
Brian R Mullen, Sydney C Weiser, Desiderio Ascencio, and James B Ackman. Automated classification of signal sources in mesoscale calcium imaging. bioRxiv, pages 2021–02, 2021.
Yina Wang, Henry Pinkard, Emaad Khwaja, Shuqin Zhou, Laura Waller, and Bo Huang. Image denoising for fluorescence microscopy by self-supervised transfer learning. bioRxiv, pages 2021–02, 2021.
Thibault Lagache, Alison Hanson, Jesús E Pérez-Ortega, Adrienne Fairhall, and Rafael Yuste. Tracking calcium dynamics from individual neurons in behaving animals. PLOS Computational Biology, 17(10): e1009432, 2021.
Adam S Charles, Nathan Cermak, Rifqi O Affan, Benjamin B Scott, Jackie Schiller, and Gal Mishne. Graft: graph filtered temporal dictionary learning for functional neural imaging. IEEE Transactions on Image Processing, 31:3509–3524, 2022.
Madhavi Tippani, Elizabeth A Pattie, Brittany A Davis, Claudia V Nguyen, Yanhong Wang, Srinidhi Rao Sripathy, Brady J Maher, Keri Martinowich, Andrew E Jaffe, and Stephanie Cerceo Page. Capture: Calcium peaktoolbox for analysis of in vitro calcium imaging data. BMC neuroscience, 23(1):71, 2022.
Leander de Kraker, Koen Seignette, Premnath Thamizharasu, Bastijn JG van den Boom, Ildefonso Ferreira Pica, Ingo Willuhn, Christiaan N Levelt, and Chris van der Togt. Specseg
is a versatile toolbox that segments neurons and neurites in chronic calcium imaging datasets based on low-frequency cross-spectral power. Cell reports methods, 2(10), 2022.
Luca Sità, Marco Brondi, Pedro Lagomarsino de Leon Roig, Sebastiano Curreli, Mariangela Panniello, Dania Vecchia, and Tommaso Fellin. A deep-learning approach for online cell identification and trace extraction in functional two-photon calcium imaging. Nature Communications, 13(1): 1529, 2022.
Honghua Guan, Dawei Li, Hyeon-cheol Park, Ang Li, Yuanlei Yue, Yung-Tian A Gau, Ming-Jun Li, Dwight E Bergles, Hui Lu, and Xingde Li. Deep-learning two-photon fiberscopy for video-rate brain imaging in freely-behaving mice. Nature communications, 13(1):1534, 2022.
Sharif Amit Kamran, Khondker Fariha Hossain, Hussein Moghnieh, Sarah Riar, Allison Bartlett, Alireza Tavakkoli, Kenton M Sanders, and Salah A Baker. New open-source software for subcellular segmentation and analysis of spatiotemporal fluorescence signals using deep learning. Iscience, 25(5), 2022.
Xinyang Li, Yixin Li, Yiliang Zhou, Jiamin Wu, Zhifeng Zhao, Jiaqi Fan, Fei Deng, Zhaofa Wu, Guihua Xiao, Jing He, et al. Real-time denoising enables high-sensitivity fluorescence time-lapse imaging beyond the shot-noise limit. Nature Biotechnology, 41(2):282–292, 2023a.
Jeffrey L Gauthier, Sue Ann Koay, Edward H Nieh, David W Tank, Jonathan W Pillow, and Adam S Charles. Detecting and correcting false transients in calcium imaging. Nature Methods, 19(4):470–478, 2022.
Andres Flores-Valle and Johannes D Seelig. Axial motion estimation
and correction for simultaneous multi-plane two-photon calcium imaging. Biomedical Optics Express, 13(4):2035–2049, 2022.
Weiyi Liu, Junxia Pan, Yuanxu Xu, Meng Wang, Hongbo Jia, Kuan Zhang, Xiaowei Chen, Xingyi Li, and Xiang Liao. Fast and accurate motion correction for two-photon ca2+ imaging in behaving mice. Frontiers in Neuroinformatics, 16:851188, 2022.
Oliver Rübel, Andrew Tritt, Ryan Ly, Benjamin K Dichter, Satrajit Ghosh, Lawrence Niu, Pamela Baker, Ivan Soltesz, Lydia Ng, Karel Svoboda, et al. The neurodata without borders ecosystem for neurophysiological data science. Elife, 11:e78362, 2022.
Zhe Chen, Garrett J Blair, Changliang Guo, Jim Zhou, Juan-Luis Romero-Sosa, Alicia Izquierdo, Peyman Golshani, Jason Cong, Daniel Aharoni, and Hugh T Blair. A hardware system for real-time decoding of in vivo calcium imaging data. Elife, 12:e78344, 2023.
Yan Zhang, Márton Rózsa, Yajie Liang, Daniel Bushey, Ziqiang Wei, Jihong Zheng, Daniel Reep, Gerard Joey Broussard, Arthur Tsang, Getahun Tsegaye, et al. Fast and sensitive gcamp calcium indicators for imaging neural populations. Nature, 615(7954):884–891, 2023a.
Zhehao Xu, Yukun Wu, Jiangheng Guan, Shanshan Liang, Junxia Pan, Meng Wang, Qianshuo Hu, Hongbo Jia, Xiaowei Chen, and Xiang Liao. Neuroseg-ii: A deep learning approach for generalized neuron segmentation in two-photon ca2+ imaging. Frontiers in Cellular Neuroscience, 17: 1127847, 2023.
Pablo Vergara, Yuteng Wang, Sakthivel Srinivasan, Yoan Cherasse, Toshie Naoi, Yuki Sugaya, Takeshi Sakurai, Masanobu Kano, and Masanori
Sakaguchi. The caliali tool for long-term tracking of neuronal population dynamics in calcium imaging. bioRxiv, pages 2023–05, 2023.
Yuanlong Zhang, Guoxun Zhang, Xiaofei Han, Jiamin Wu, Ziwei Li, Xinyang Li, Guihua Xiao, Hao Xie, Lu Fang, and Qionghai Dai. Rapid detection of neurons in widefield calcium imaging datasets after training with synthetic data. Nature Methods, 20(5):747–754, 2023b.
Jacopo Bonato, Sebastiano Curreli, Sara Romanzi, Stefano Panzeri, and Tommaso Fellin. Astra: a deep learning algorithm for fast semantic segmentation of large-scale astrocytic networks. bioRxiv, pages 2023–05, 2023.
Xinyang Li, Xiaowan Hu, Xingye Chen, Jiaqi Fan, Zhifeng Zhao, Jiamin Wu, Haoqian Wang, and Qionghai Dai. Spatial redundancy transformer for self-supervised fluorescence image denoising. bioRxiv, pages 2023–06, 2023b.
Junmo Cho, Seungjae Han, Eun-Seo Cho, Kijung Shin, and Young-Gyu Yoon. Robust and efficient alignment of calcium imaging data through simultaneous low rank and sparse decomposition. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pages 1939–1948, 2023.
Biafra Ahanonu, Andrew Crowther, Artur Kania, Mariela Rosa Casillas, and Allan Basbaum. Long-term optical imaging of the spinal cord in awake, behaving animals. bioRxiv, pages 2023–05, 2023.