-
Notifications
You must be signed in to change notification settings - Fork 10
User Manual
We provide a step-by-step user manual here. The user needs to install python3.8.
Please referfor instructions
cd PatchSorter
python PS.py
http://localhost:5555
The user could change the port number in the config.ini file.

- PatchSorter requires users to upload images and masks, supported types of masks are QuickAnnotator Masks, Binary Mask, Labelled Mask and Indexed Mask.
- User could drag drop images or could provide a folder path to upload from, or provide a comma seperated list of image and mask names with folder path.

The uploaded images are divided in PS in patches of a user defined size, currently the User Interface supports patches of 32px, 64px and 256 px, this needs to be configured in config.ini file prior to Make Patches step.
Refer to --https://github.com/choosehappy/PatchSorter/wiki/Hyper-Parameter
The image below shows, make patches successfully completed, and the next step is View Embed which directs user to Embedding Page.

The image below shows, training DL successfully completed, and the next step is Embed Patches

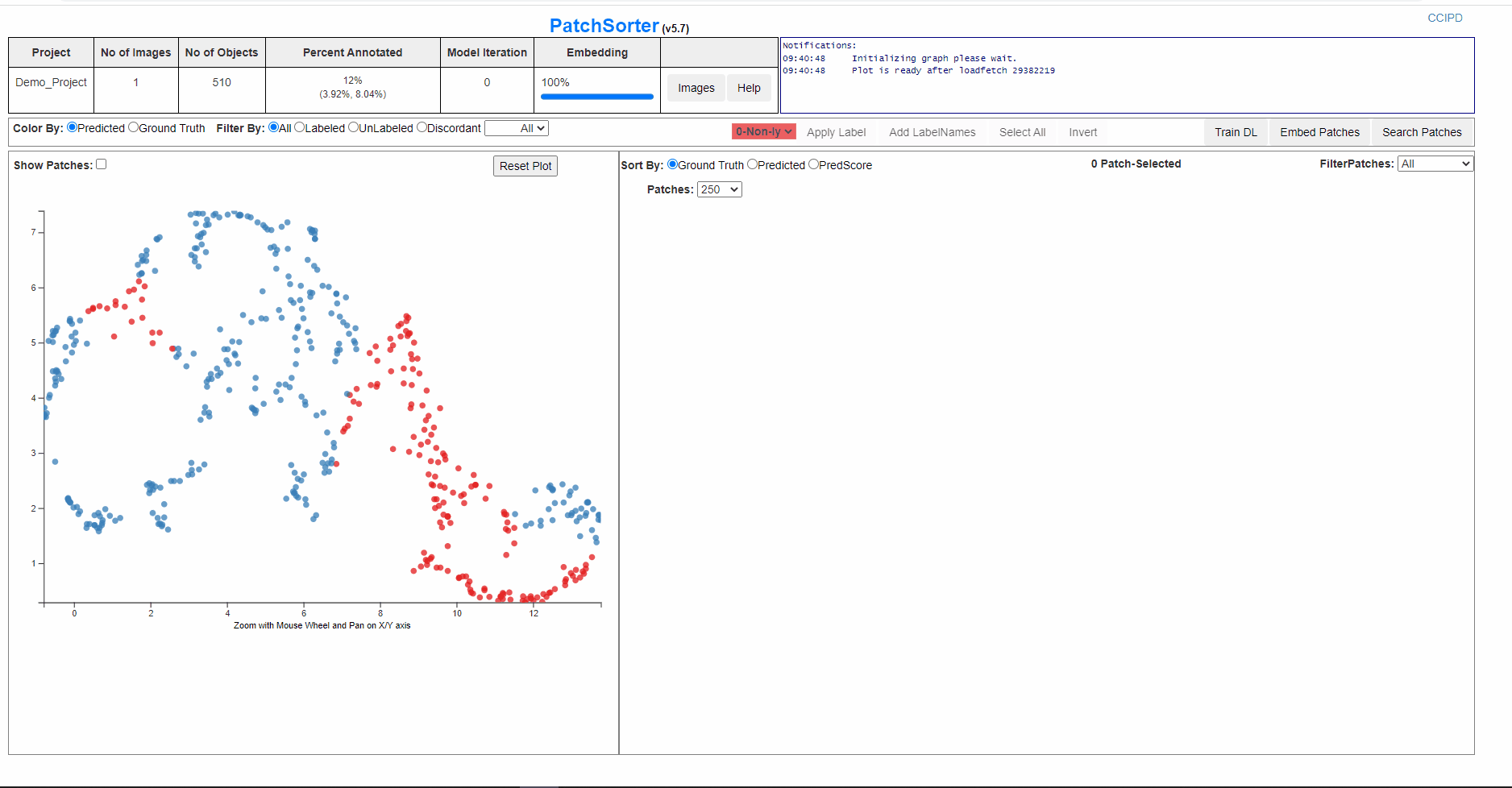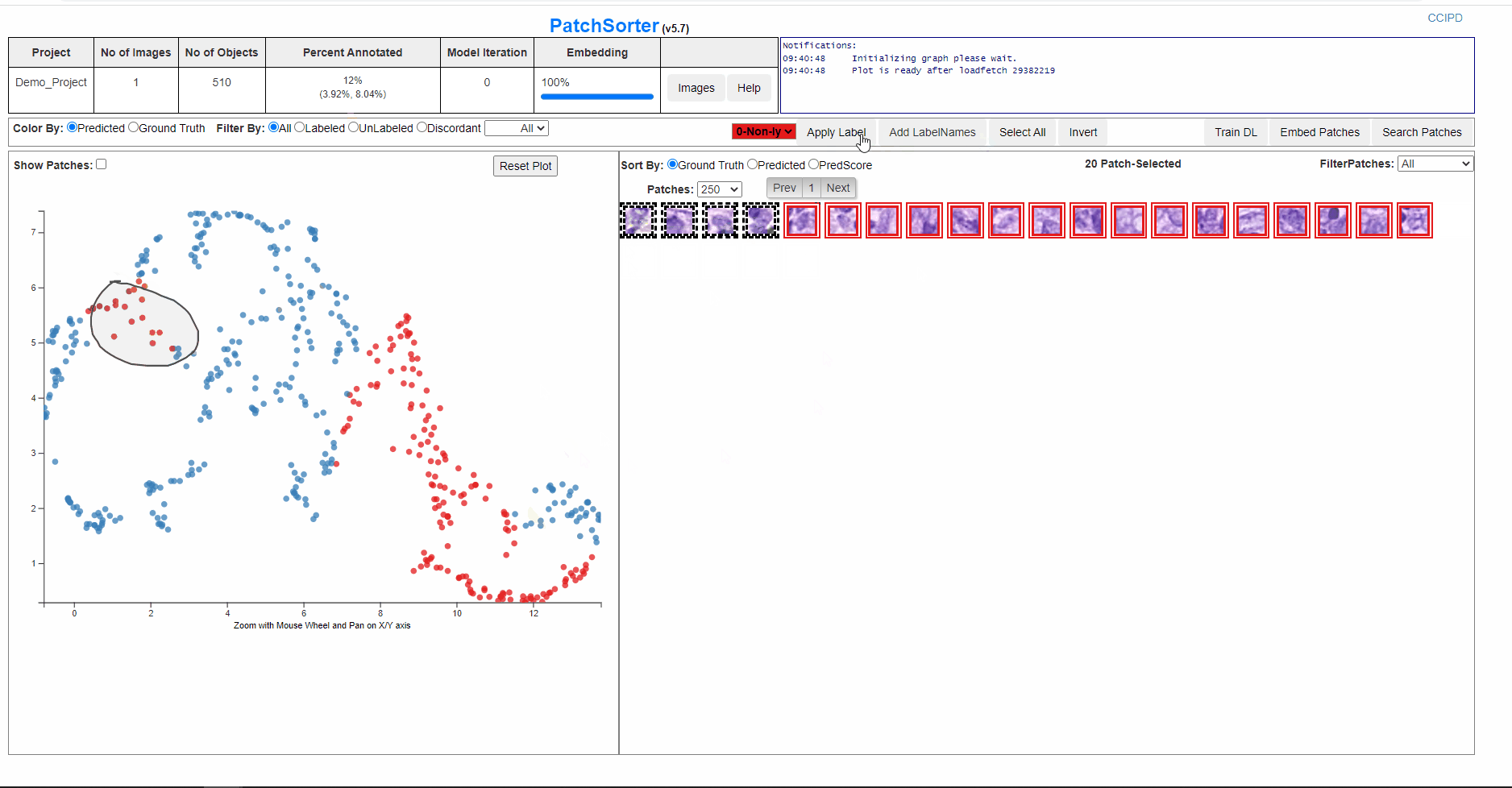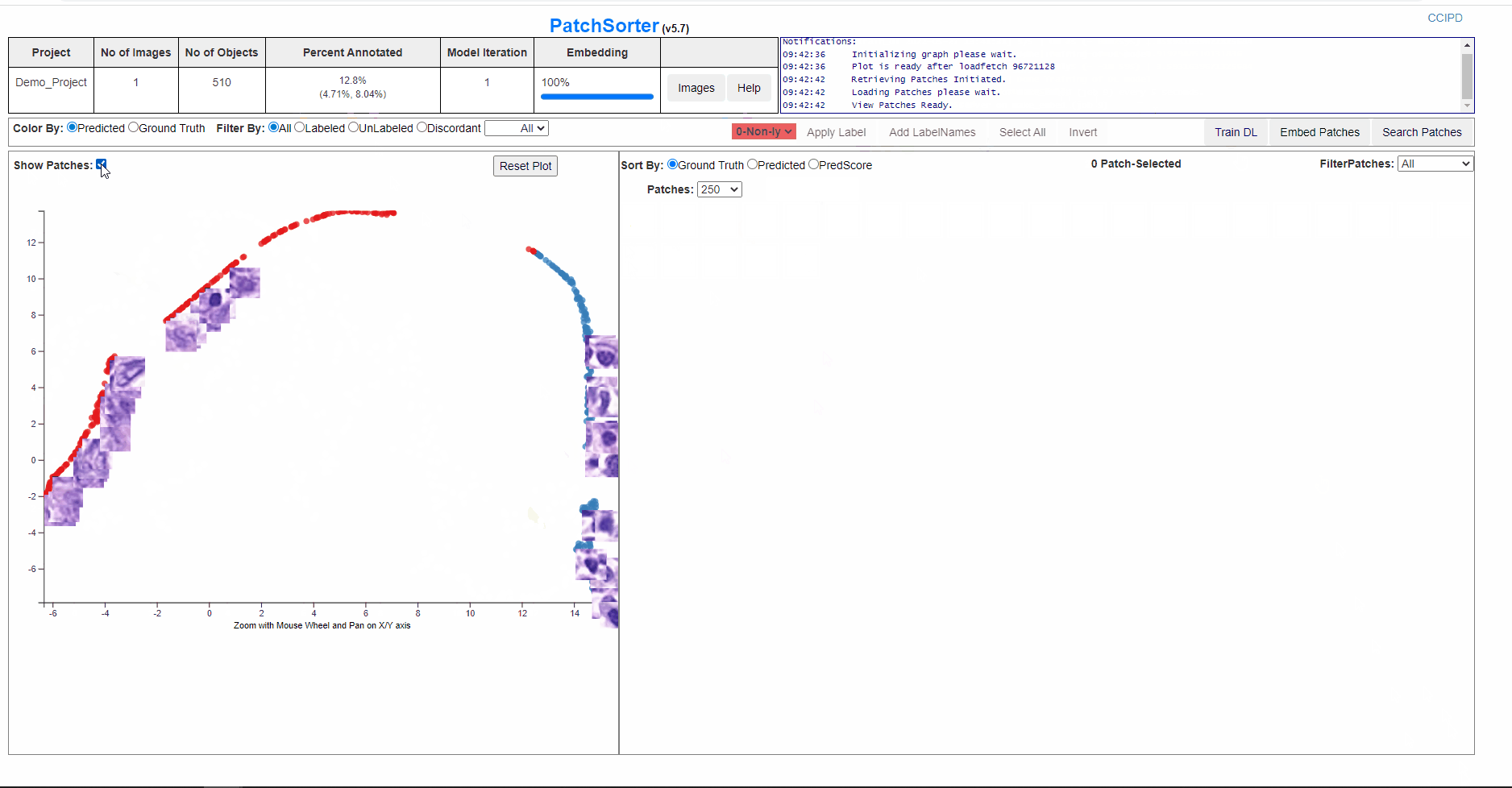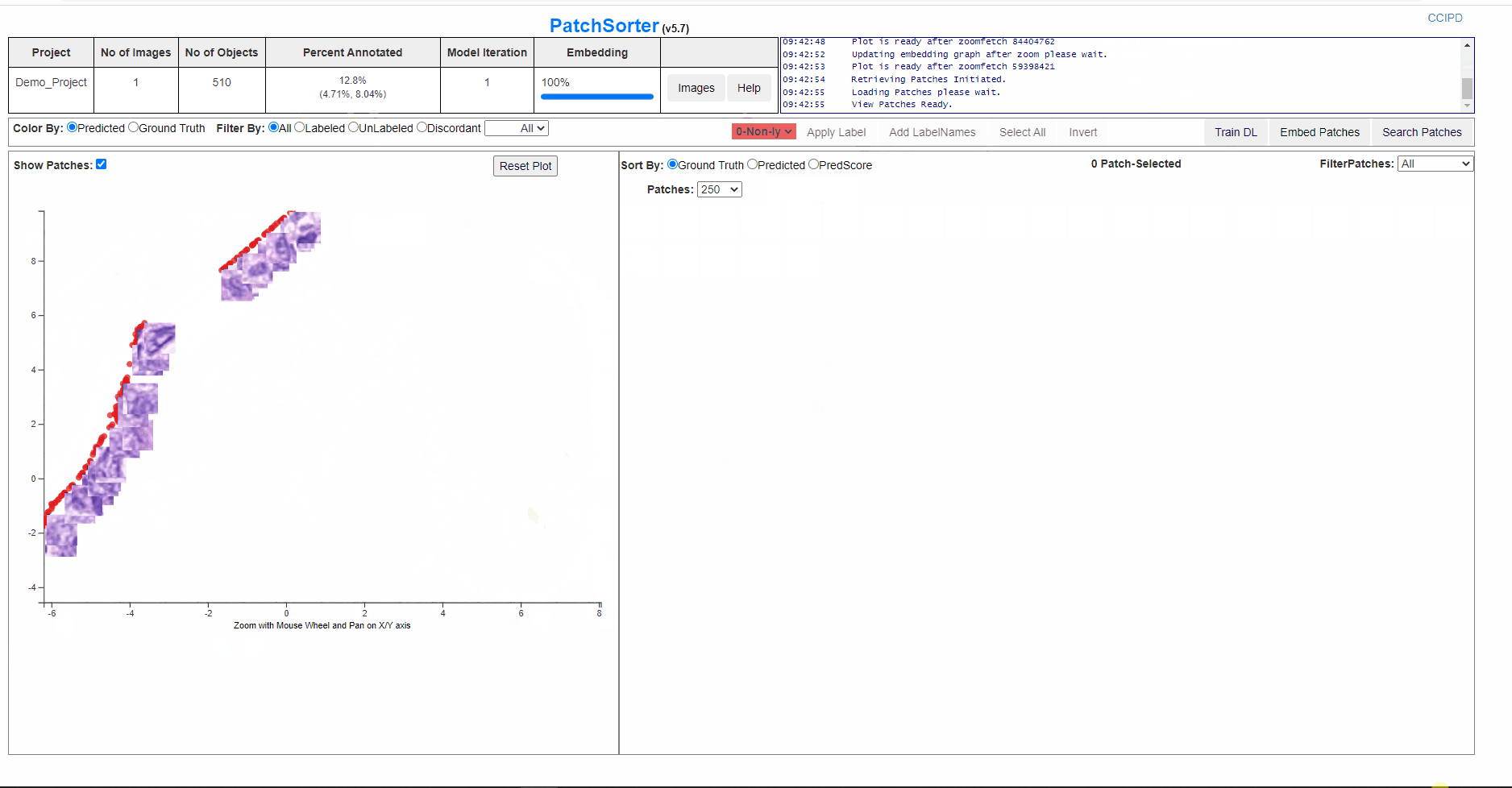
After the patches are embedded, a 2D scatter plot with all the patches represented as dot will be presented to the user. The patches will be clustered based on unsupervised Deep Learning Model and with further labelling , re training and embedding they will be separated into homogenous clusters.
User can click on Show Patches to check the distribution of the patches on the plot.

On lasso over the plot points, the patches are loaded in the grid form.



The screen below is showing process of re-training and embedding. And the plot will get refreshed with the clusters seperated better and would be effective for a faster labelling.
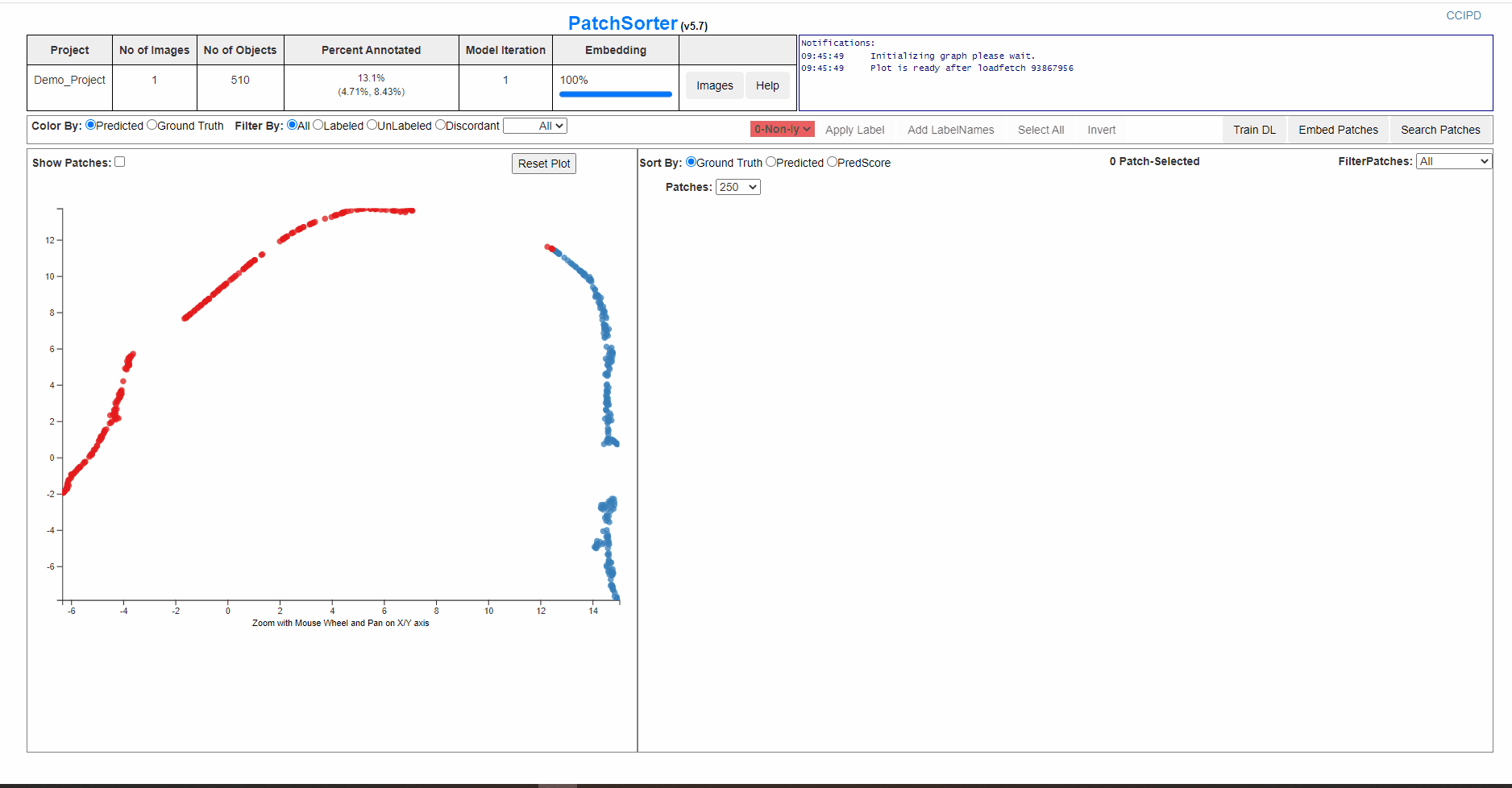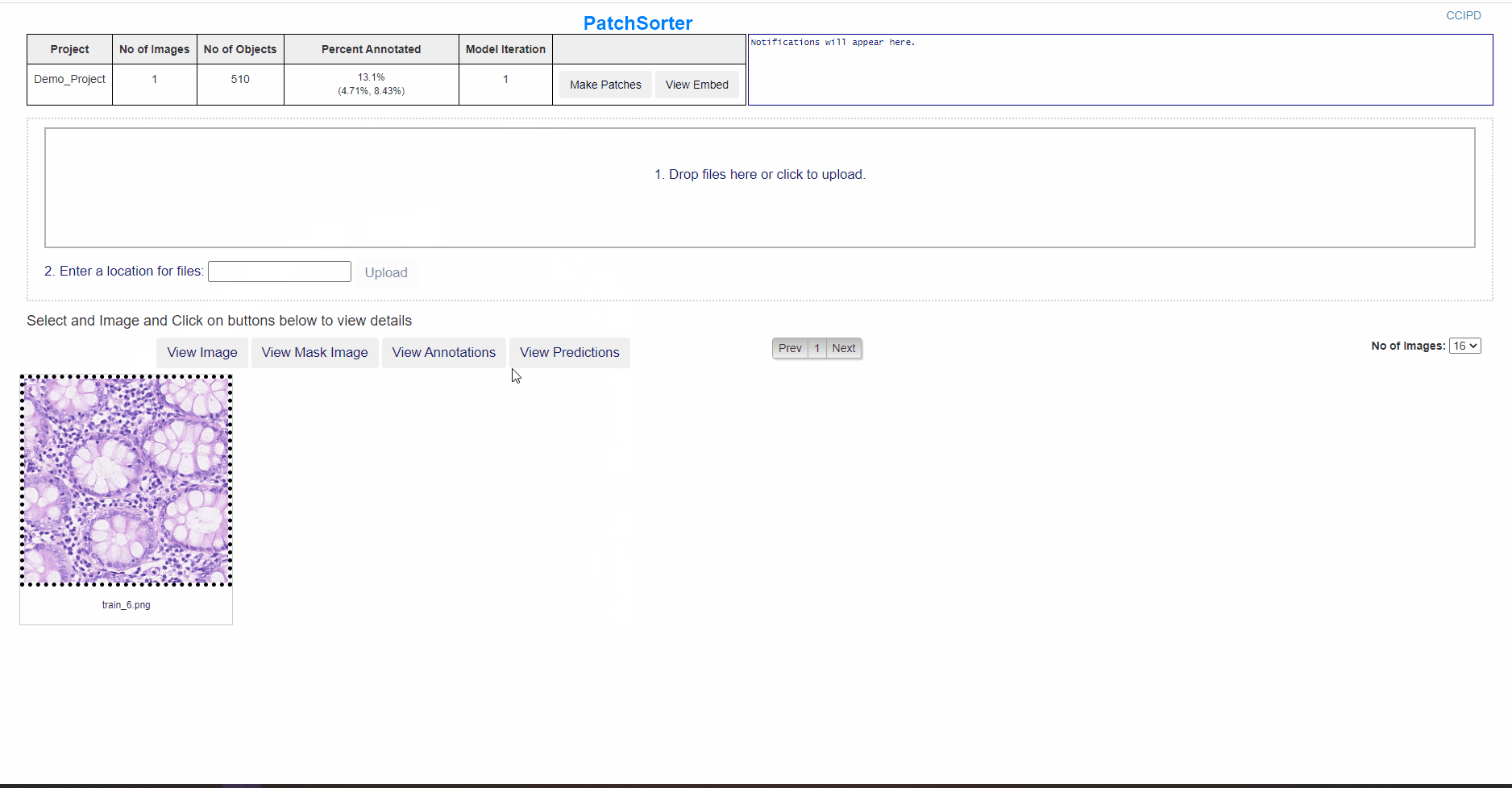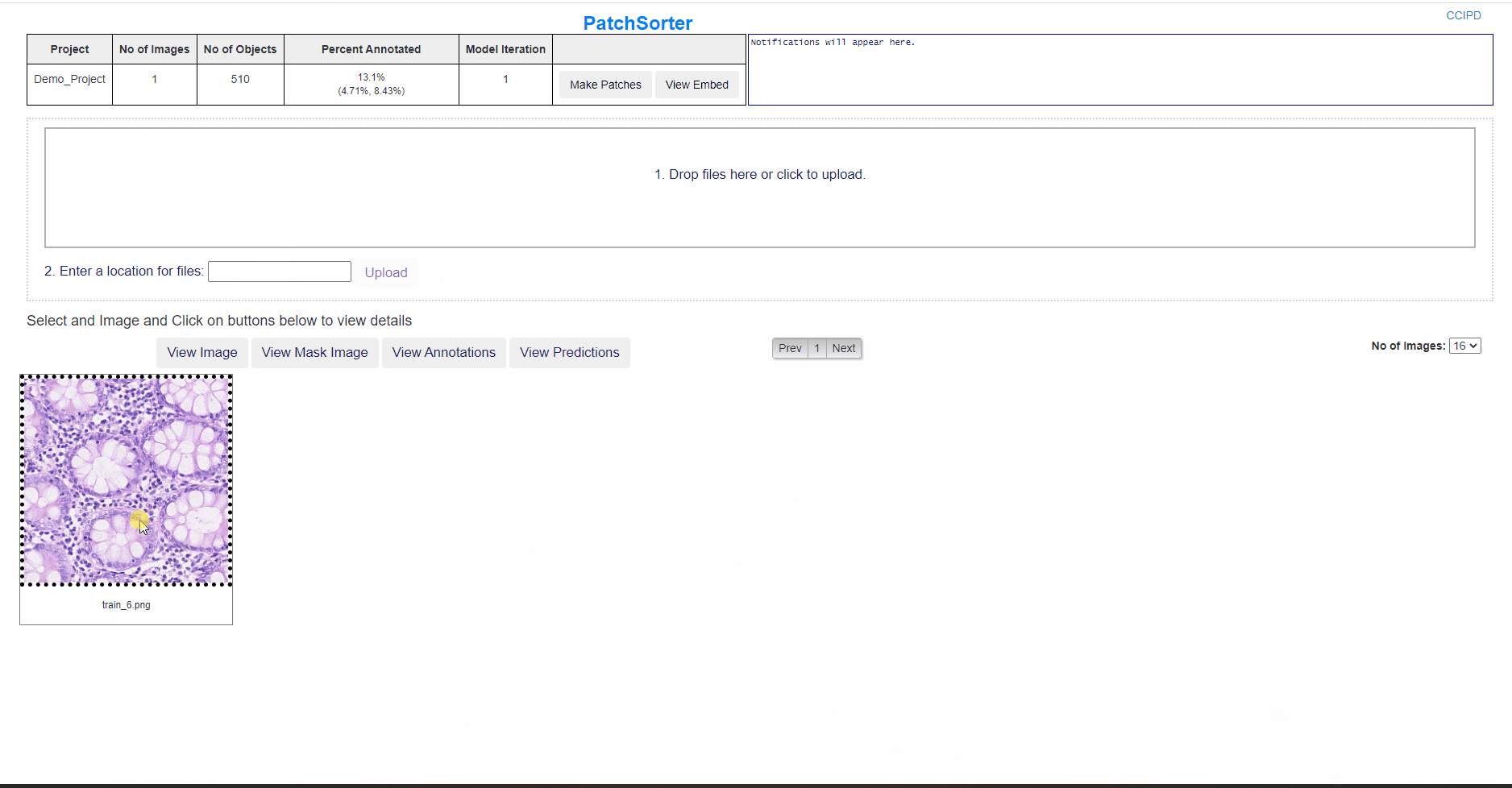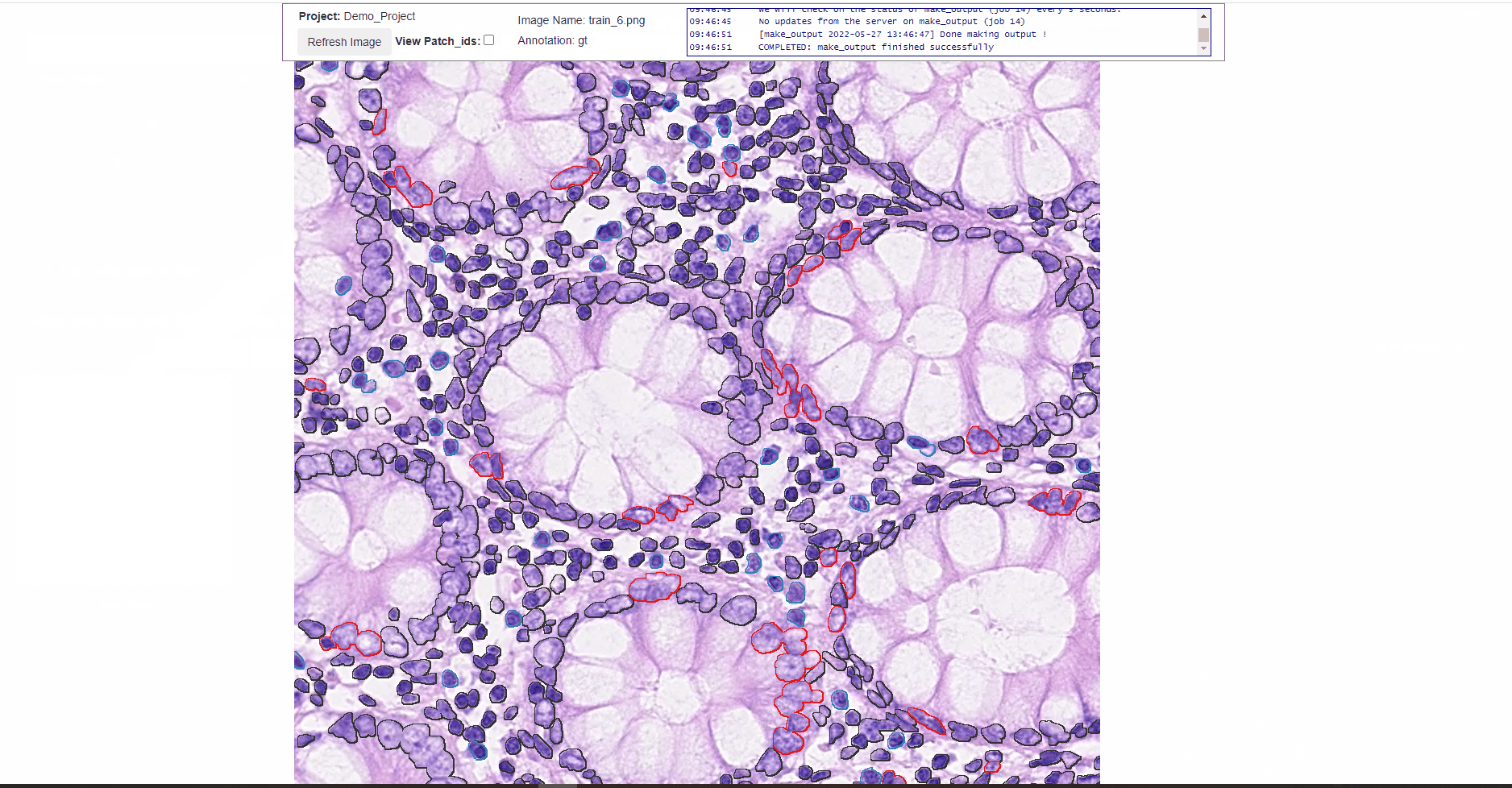
- Viewing the DL learning model predicted cell/object labels overlayed on the image.
- Viewing the annotated cell/object labels (labels that user assigned) overlayed on the image. Black color here would depict unlabeled data. These features help the user to view the labels with a better context and has an option to fine tune the labelling and update them back in the system.
PS's Wiki is complete documentation that explains to user how to use this tool and the reasons behind. Here is the catalogue for PS's wiki page:
Home: